Back to Journals » Journal of Inflammation Research » Volume 15
Insights into the Role of Commensal-Specific T Cells in Intestinal Inflammation
Authors Gehlhaar A , Inala A, Llivichuzhca-Loja D, Silva TN, Adegboye CY, O'Connell AE, Konnikova L
Received 24 November 2021
Accepted for publication 19 February 2022
Published 14 March 2022 Volume 2022:15 Pages 1873—1887
DOI https://doi.org/10.2147/JIR.S288288
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Arne Gehlhaar,1,* Ashwin Inala,1,* Dhana Llivichuzhca-Loja,1,* Tatiana N Silva,1,* Comfort Y Adegboye,2 Amy E O’Connell,2,3 Liza Konnikova1,4,5
1Department of Pediatrics, Yale University, New Haven, CT, USA; 2Division of Newborn Medicine, Boston Children’s Hospital, Boston, MA, USA; 3Department of Pediatrics, Harvard Medical School, Boston, MA, USA; 4Department of Obstetrics, Gynecology and Reproductive Sciences, Yale University, New Haven, CT, USA; 5Program in Human and Translational Immunology, Yale University, New Haven, CT, USA
*These authors contributed equally to this work
Correspondence: Amy E O’Connell, Division of Newborn Medicine, Boston Children’s Hospital, 300 Longwood Ave, Boston, MA, USA, Tel +1 617 919-2358, Email [email protected] Liza Konnikova, Department of Pediatrics, Yale University, 375 Congress Ave, LSOG 405B, New Haven, CT, USA, Tel +1 203 737-8016, Email [email protected]
Abstract: Trillions of microorganisms exist in the human intestine as commensals and contribute to homeostasis through their interactions with the immune system. In this review, we use previous evidence from published papers to elucidate the involvement of commensal-specific T cells (CSTCs) in regulating intestinal inflammatory responses. CSTCs are generated centrally in the thymus or peripherally at mucosal interfaces and present as CD4+ or CD8+ T cells. Bacteria, fungi, and even viruses act commensally with humans, warranting consideration of CSTCs in this critical relationship. Dysregulation of this immunological balance can result in both intestinal inflammation or damaging autoimmune responses elsewhere in the body. Given the relative novelty of CSTCs in the literature, we aim to introduce the importance of their role in maintaining immune homeostasis at barrier sites such as the intestine.
Keywords: commensal, T cells, immunity, microbiome
Introduction
Over the past 15 years, there has been an increasing interest in host–microbiome interactions and their involvement in the development of immunity and disease. Commensal microbiota are those that commonly reside on barrier sites such as the gut and do not initiate disease under normal conditions.1 Commensals in our bodies express antigens that can be detected by the organism’s immune system and give rise to commensal-specific lymphocytes.2,3 This review will focus on commensal-specific T cells (CSTCs) as those that express an αβ T cell receptor (TCRs) and have a moderate to high affinity for commensal-derived peptides presented on major histocompatibility complex (MHC) class I or II molecules. The results discussed in this review often do not focus solely on the TCR mediated functions, but rather focus on observed changes in T cell populations or functions, which, in addition to TCR specific effects, may result from non-TCR specific functions of commensal organisms. Nonetheless, given the limited literature that currently exists on CTSCs, it is beneficial to include non-TCR specific findings as these data might hint at TCR specific CSTC functions. CSTCs not only contribute to defense against microbiota by inducing barrier-protective cytokines,4 but also play important roles in immune development, T-cell differentiation and immune responses at these barrier sites between the external environment and internal organs.3
There are trillions of microbiota present in the human intestine. The presence of these bacteria, fungi, or even viruses prompts consideration of the specific mechanisms the immune system uses to maintain gut homeostasis and prevent pathogenicity. Specifically, many studies have sought to better understand how the numerous T cell subtypes, such as CD4+ and CD8+ T cells, respond to these commensal organisms. This review will therefore explore the generation of CSTCs, how they interact with key microbial organisms in the gut, and how this connects to disease.
Generation of Commensal-Specific T Cells
T cells are classically characterized in subsets based on co-receptor molecule expression and transcriptional or cytokine profiles. CD4+ T cells, classically described as helper T (Th) cells for their ability to generate cytokines and propagate an immune response, can be further subclassified based on the cytokine or transcription factors they produce, as previously covered by many comprehensive reviews.5,6 CD8+ T cells, or cytotoxic T cells, have a primary role in directly killing infected, injured, or metastatic cells.7 The CD4 and CD8 subgroups are further subclassified into effector and memory cells based on expression of homing and activation-related ligands.8 There are important differences in commensal T cell properties between T cell subgroups, with commensal interactions leading to expansion of some subgroups and inhibition of others, as described below.
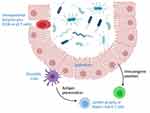
CSTCs can be generated centrally, in the thymus, and peripherally at the mucosal interfaces where commensal bacteria reside. Within the thymus, selection of T cells specific for commensals occurs after trafficking of commensal antigens from the intestine to the thymus by intestinal dendritic cells.9,10 Most of the CSTCs in the thymus are positively selected CD4+ T cells, again highlighting the critical role of antigen trafficking in driving positive selection.10 The generation of thymic CSTCs is dependent on the type of commensal microorganism present in the microbiota, such as segmented filamentous bacteria (SFB). For example, the colonization of young B6 mice with SFB resulted in an expansion in SFB-specific CD4+ T cells which was not observed in young SFB-negative mice.10 Similar results were found in mice colonized with Escherichia coli.10 Bacterial DNA from SFB and other commensal organisms has been detected in trafficking specifically to the mesenteric lymph nodes (MLN) and the thymus, with a peak detection around one week after intestinal colonization, which occurred in young mice (freshly weaned) and not in mature mice (12 weeks).10 CX3CR1+ dendritic cells (DCs) are the primary antigen presenting cells that expand in the thymus after introduction of commensal bacteria.10 A separate population of DCs, CCR9+ plasmacytoid dendritic cells (pDCs) also migrate to the thymic medulla from the periphery and present antigens to specific thymocytes to induce tolerance, and these cells have been postulated to function in tolerance induction to commensals, but evidence for this remains elusive.11,12
Locally in the intestine, two DC subtypes (CD103+, CX3CR1+) also play a critical role in responses to commensals, with CD103+ as the dominant subtype.13 CD103+ DCs drive the proliferation of commensal-specific CD4+ and CD8+ T cells by presenting commensal antigens in mouse intestines.14 Trafficking of CX3CR1+ DCs to the MLN has also been shown to be regulated by commensal bacteria,5 although the downstream impacts of CX3CR1+ DCs in the gut are less clearly understood.
Intestinal epithelial cells (IECs), while not traditionally considered immune cells, are critical in immunologic responses to commensal bacteria.6 Functions of IECs are influenced by interactions with commensals and pathogenic bacteria, including proliferation,7 mucus production,15,16 expression of Toll-like receptors and inflammasome components,17–19 and production of antimicrobial peptides (AMP) including defensins, cathelicidins, and RegIII isoforms.20,21 IECs interactions with commensal bacteria seem to be particularly critical for induction of Th17 cells, discussed in more detail below.
Several regions of the intestine harbor important T cell populations. The submucosal immune organs, including Peyer’s patches and lamina propria feature mostly CD4+ T cells. In contrast, the intestinal epithelial lymphocyte (IEL) population contains higher proportions of γδ T cells and CD8+ T cells. Because of the proximity of IELs to the gut lumen, these cells often directly interact with commensal bacteria. T cells in the secondary lymph tissues rely on antigen presentation of commensals by DCs or secondary signals propagated by DCs or IECs as outlined above (Figure 1).
CD4 Commensal T Cells
Under homeostatic conditions, commensal and host signals provide cues to naive CD4+ T cells to induce their differentiation into various effector subsets. In the intestines, CD4+ T cells are expanded by the presence of commensal bacteria. Memory CTSCs were found both in circulation and in the tissues. Moreover, they were heterogeneous in their function producing IL-17, IL-10, IFN-γ and TNF-α.4 On the contrary, infection and inflammation drive commensal-specific CD4+ T cells towards Th1 and Th17 differentiation.9
Th1 Cells
Microbial colonization directly enhances Th1 CD4+ T cell gene responses in the intestine, including STAT1, IFN-γ and IL-18.22 A study by Fiona Powrie’s group demonstrated that, in humans, commensal bacteria specific CD4+ T cells (those reactive to E. coli, L. acidophilus, Bifidobacterium, F. prausnitzii, B. vulgautus, R. intestinalis, R. obeum, S. typhimurium and C. difficile) produce Th1 cytokines including IFN-γ and TNF-α, among others.4 In contrast to other Th populations in the gut that appear to promote homeostasis with commensals, commensally regulated Th1 populations are linked to pathology and disease. The spontaneous colitis mouse model, C3H/HeJBir, is linked to spontaneous production of inflammatory Th1 populations after exposure to enteric bacteria.23 Whether this process is mediated by CSTC receptors is not known. CBir1 transgenic mice with a TCR specific for an antigen on microbial flagella also develop spontaneous colitis with a predominant Th1 cytokine signature (IL-1b, TNF-α, IL-6).24 Indeed, Th1 cell subsets are thought to be the primary drivers of pathology in inflammatory bowel diseases, conditions that are known to have associated changes in intestinal microflora.25
Th2 Cells
Th2 populations are important in allergic immune responses and are dominated by the production of IL-4, IL-10 and IL-13.26 Th2 CSTCs have not been described at the time of this review, but commensal-specific responses play an important part in regulation of the Th2 compartment. In contrast to Th1 and Th17 cells, commensal bacteria appear to inhibit production of Th2. Commensal bacteria trigger production of soluble CD14 (sCD14) from monocytes and both sCD14 and sCD83 from dendritic cells, which prevented allergic sensitization and expansion of Th2 cells in neonatal blood.27 Production of transforming growth factor-beta (TGF-β) by other CD4+ T cells in response to commensal bacteria has also been shown to play a role in suppressing Th2 cell development.28
Th17 Cells
Th17 cells are distinctly important for surveillance at barrier sites, including the intestine, and intestinal microbial stimulation directly stimulates Th17 T cell production.22 Some members of the microbiota adhere to epithelial cells in the intestine, causing induction of serum amyloid A expression (SAA).29 SAA production enhances Th17 cell specification in dendritic cell co-culture experiments in an antigen-specific manner, suggesting that the epithelial cells produce signals that influence CD4 T cell profiles when antigen presenting cells interact with T cells in the MLN.29,30 SFB, in particular, play an important role in stimulating commensal Th17 production in the intestine.22,31,32 SFB stimulate formation of germinal centers in Peyer’s patches and can separately induce generation of tertiary lymphoid tissue in the intestine, and these lymphoid centers were significantly enriched for Th17 cells.32 Th17 cell generation in response to commensals is regulated in part by type 3 innate lymphoid cells (ILC3), which can deplete commensal specific CD4+ T cells in the gut.33
Th17 cells also play a critical role in fungal-specific immunity. The clinical importance of intestinal Th17 cells in commensal fungal interactions is highlighted by the fact that human mucocutaneous candidiasis is related to gene defects which cause deficiency Th17 cell production.34,35 IECs are critical in Th17 cell induction, and when SFB interactions with IEC are interrupted, Th17 cells are not induced,29 indicating that IECs are critical mediators in the Th17 response to SFB and other adherent microbes. In turn, production of AMPs by IECs is strongly influenced by IL-17 and IL-22 production by Th17 cells, suggesting a feedback loop.36
Regulatory T Cells
Regulatory T cells (Tregs) can also be dominantly induced from naïve T cells by commensal bacteria in the periphery, specifically the distal MLN, in response to commensal antigens.37–39 This subset of Tregs is typically referred to as “inducible” Tregs.37 Generation of peripheral Tregs is driven by direct and indirect signals from commensal bacteria such as Clostridium species and B. fragilis.38,40,41 In the colon, Clostridium species induce the secretion of TGF-β by intestinal epithelial cells and indoleamine-2,3-dioxygenase by intestinal epithelial cells and dendritic cells, which collectively favor the differentiation of Tregs.40,42 Polysaccharide A (PSA) from B. fragilis also induces IL-10 secretion from commensal-specific Tregs in the lamina propria.42,43 CD103+ DCs travel from the lamina propria to the mesenteric lymph node and present commensal antigens to CSTCs as described above,13,44 leading to the production of TGF-β and conversion of retinol into retinoic acid.9,14 Retinoic acid and TGF-β are partly responsible for the differentiation of commensal-specific Tregs.9,13 The increased expression of CD25, CD5 and cytotoxic T lymphocyte antigen 4 (CTLA-4) signals during TCR stimulation contribute to the induction of Foxp3 that specifies Treg cell lineage.45 Short chain fatty acids (SCFAs), a dietary metabolite produced by gut bacteria, increase the differentiation of Tregs, the stability of Foxp3 gene expression, and the expression of IL-10 in Tregs.41,42 Foxp3 expression is upregulated when gnotobiotic mice are transplanted with fecal microbiota from a conventional mouse,22 again highlighting the critical role of commensals in expansion of this population.
CD8 Commensal T Cells
CD8+ T cells expressing the α and β T cell receptor subunits (CD8αβ+) constitute the majority of IELs. Similar to Th17 cells, CD8αβ+ populations are significantly diminished in germ-free (GF) mice, indicating that commensal bacteria drive expansion of this population.46 This commensal-specific expansion is dependent on TLR signaling, as the CD8αβ+ IEL population is diminished in TLR2−/−, TL4−/− and TLR9−/− mice. Furthermore, stimulation of CD8αβ+ IELs by commensals lead to potent production of defensins, which are important in intestinal defenses against pathogenic bacteria and viruses, and IFN-γ.46–48 The importance of the microbiome in regulation of the CD8+ T cell IEL compartment is highlighted by the finding that dysbiosis leads to exhaustion of IFN-γ-producing CD8+ T cells in the intestinal microenvironment, increasing susceptibility to tumorigenesis.49 CD8αβ+ IELs express CD44 (activation marker) and T-bet (lineage marker), indicating that they arise from conventional CD8+ T cells that originate in the thymus.48,50 While the exact circumstances of how these T cells come to reside within the epithelium are not yet fully understood, it is probable that circulating CD8+ T cells with a commensal-specific antigen receptor become stimulated as they circulate through the intestinal lymph or submucosal lymph tissue, and this activation results in expression of tissue homing receptors, such as CD103 which is known to be expressed on the CD8αβ+ IELs.48
Key Microbes Driving CSTC Population
Bacteria
Lactobacillus
While known for their beneficial probiotic effects in fermented foods, Lactobacilli engage in a commensal relationship with humans in various ways.51 Lactobacilli are Gram-positive, anaerobic, rod-shaped bacteria that do not form spores. In humans, prenatal vertical transmission of microbiota has recently been explored. Lactobacillus is the predominant bacteria in the vagina, where it regulates pH and produces lactic acid to prevent infection.52 Therefore, infants born vaginally are colonized with Lactobacilli at birth, unlike those born by Caesarean section (C-section), who mainly acquire maternal microbes found on the skin, such as Staphylococcus.53 As Lactobacillus occurs naturally in the gut microbiota by its acquisition at birth, further consideration of its commensal relationship with the immune system is necessary.
With this in mind, specific immune responses to Lactobacilli most likely occur after birth. There is evidence that both circulating and gut-resident human CD4+ T cells are responsive to L. acidophilus in healthy human adults.4 Like other types of memory cells, CD4+ T cells that are reactive to Lactobacilli and other microbiota express adhesion molecules and chemokine receptors (CCR) that can promote entry into the intestine under inflammatory conditions.4 In L. acidophilus, CD4+ T cells had expression of CCR2, CCR4, and CCR7 >50%, which direct access to the intestinal mucosa and CSTCs are about fivefold more prevalent in the mucosa than in the periphery.4 Moreover, a large proportion of CSTCs in this study expressed CCR6 and CD161, markers enriched in Th17 and Tregs.4 CSTC produce IL-17, IFN-γ and IL-10 upon stimulation suggesting a Th17/Th1/Treg predominance.4 These findings highlight the complex balance that must exist between Lactobacilli and their CSTCs in maintaining tolerance in the intestine. Moreover, this commensal relationship begs the question of how Lactobacillus, among other microbiota, can contribute to gut pathologies like IBD.
Clostridium
Although certain species of Clostridium, such as C. perfringens and C. tetani, are commonly known for their pathogenic effects, the bacteria act mainly as a commensal with humans. Clostridia are Gram-positive, rod-shaped bacteria that form endospores, which provide them with a survival advantage under harsh environmental conditions.54 Clostridium colonizes during infancy in humans via maternal transmission of breastmilk and occupies the interfold area of the intestinal mucosa that allows for enhanced communication with intestinal cells throughout life.54 This close proximity facilitates immune processes in the intestine, such as by stimulating αβ TCR IELs in the large intestine.54 Umesaki et al found that the ratio of CD8+ to CD4+ cells in αβ IELs was significantly higher in the large intestines of Clostridium colonized mice compared to those colonized with SFB mice, suggesting the importance of the Clostridium species on the CD8/CD4 balance.55 Moreover, other evidence suggests that Clostridium impacts CD4 differentiation, where GF mice colonized with a commensal Clostridium subtype had an accumulation of colonic CD4+ Tregs expressing Foxp3 in the lamina propria (LP), whereas specific-pathogen free (SPF) mice or those treated with Lactobacillus did not.40 Similar findings have been reported in studies involving Clostridium native to the human microbiota.56 By isolating 17 strains from human microbiota, Atarshi et al 2013 found that Clostridium can elicit a cytokine response through the production of short-chain fatty acids that induce TGF-β, which contributes to the specific development of colonic Tregs in mice.54,55 It has been proposed that intestinal epithelial cells communicate closely with immune cells and drive this progression by secreting cytokines such as IL-7 to regulate IELs.54,57 Collectively, these findings emphasize the potential specificity of Clostridium in inducing colonic Treg expression and prompt further consideration of how its dysregulation in the human microbiome can contribute to intestinal disease.
Escherichia coli
Escherichia coli (E. coli) is a ubiquitous bacterial species in the mammalian intestine and remains one of the most utilized model organisms of medical and scientific research. The Gram-negative, facultative anaerobic, rod-shaped58 bacteria typically colonize human lower intestines during the first days of life via vaginal delivery and maternal transmission of breastmilk59–61 and provide important colonization protection against pathogenic microorganisms.62 The role of this colonization protection is rather significant for a commensal as widespread as E. coli, but due to its high genetic plasticity, different strains also acquired pathogenic properties,63 a trait that, in combination with frequent antibiotic resistances,64 classifies those E. coli strains as dangerous pathogens.
With regard to CSTCs, two particular mechanisms are apparent, both related to the E. coli flagellum. It has been shown that an antigen on flagella was able to induce proliferation of Th1 cells in the human intestine, although via an indirect mechanism of IL-6 production by DCs.27 It has yet to be determined with certainty, whether this innate mechanism specifically targets commensal or flagella specific T cells.
In addition, E. coli has been shown to induce Treg expansion in a manner specific to the intracellular location of the antigen presented by the E. coli strain. Intestinal Tregs have been shown to regulate the amount of IgA positive B cells in the mouse gut, leading to higher intestinal IgA production, which is linked to reduced uptake of luminal flagellin, contributing to gut homeostasis.65 E. coli specific Tregs also show reduced IL-10 production in Crohn’s disease, highlighting their importance for gut homeostasis in humans.66
These studies suggest that E. coli exhibits a variety of protective functions, from broad unspecific such as colonization protection to rather specific ones linked among others to the flagella of motile strains. Additional experiments are required to address the specific function of E. coli specific T cells subpopulations in the human intestine.
Fungi
Candida albicans
Historically, viruses and bacteria have been the focus of microbe/host interactions, mostly due to their clinical relevance when compared to fungi. However, over the past decade, there has been an increased interest in the function both in health and disease of commensal fungi in the human microbiota,67 particularly in the intestine.
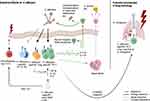
One of the most prevalent intestinal commensals, both in frequency in the population68,69 and in the literature, is Candida albicans. The interest surrounding this commensal stems not only from its frequent use as a model organism, but also its clinical significance and central role for antifungal immunity.70
C. albicans is a dimorphic fungus, existing either as a haploid or diploid strain. Colonization occurs orally, for 22–24% of all infants sub partus;71 in general, about 40–60% of patients are colonized with C. albicans under normal conditions.62 The early colonization of the intestine64 and skin,72 combined with the ability of neonatal T cells to mount an efficient response to C. albicans73 might provide a possible explanation for the high prevalence of C. albicans specific Th17 cells in humans. Fungi in general, and especially C. albicans, are of particular interest for this review, as gut-resident CD4+ T cells were shown to have a larger percentage of Th17 differentiated cells when compared to blood-circulating central memory T cells (Tcm) in humans,4 an effect pronounced in IBD. This is relevant, because Th17 cells are generally considered the main adaptive driver of antifungal immune mechanisms and, consistently, C. albicans has been shown to be the strongest known inducer as well as a target of Th17 responses (Figure 2).74,75 Gut-resident T cells are enriched in their reactivity for C. albicans, about 1500–2000 in 106 gut-resident CD4+ are C. albicans specific, predominantly of the memory phenotype.4 The differentiation of antifungal Th17 into blood circulating central memory Tcm has also been reported in humans,70,74 ensuring central whole organism protection against fungi infection and overgrowth. Apart from Th17 cells, C. albicans has also been shown to induce Foxp3+ Tregs, with fungal-specific Tregs being highly expanded in human peripheral blood.76 It has been speculated that the anti-fungal Tregs might benefit Th17 differentiation by depriving activated, classical T cells of IL-2,77,78 although Th17 immunity as well as fungal colonization were not affected in the absence of Tregs in mice.79 Therefore, the role of fungi-specific Tregs remains largely unclear. Small proportions of anti-fungal Th1 and Th2 cells have also been observed, their potential functions in respect to gut fungi remain largely unknown. A mechanism of potential cross reactivity with A. fumigatus in the lung, a known trigger of Th2 responses, has been suggested in mouse models.80
C. albicans is the predominant inducer of human Th17 responses, while Th17 cells directed against other fungi (Pityrosporum orbiculare,81 Hormodendrum strains, A. fumigatus,70 other Candida strains,82 etc.83) are induced by cross-reactivity to C. albicans (Figure 2).70 Interestingly, the mechanism appears to be rather permissive, neither being restricted to one barrier site nor to a confined group of microbes. C. albicans specific and cross reactive Th17 cells have been shown to switch to an activated state and expand in a selective manner in patients with acute allergic bronchopulmonary aspergillosis and other types of airway inflammation.70 Similarly, it has been proposed that the early C. albicans specific, skin resident Th17 subset serve as a reservoir for cross-reactive Th17 cells upon Malassezia species infection in the skin,77 another fungal commensal.
The influence of C. albicans specific Th17 cells spans beyond fungal recognition, as it has been suggested, that these T cells might modulate the neutrophil-mediated immune response against Staphylococcus aureus.77 S. aureus itself is the second strongest inducer of human Th17 responses,84 suggesting a possibility of mutual influence of C. albicans and S. aureus on IL-17 secretion. However, there are some differences as C. albicans specific Th17 co-secrete IFN-γ, while S. aureus specific Th17 showed IL-10 secretion upon restimulation.84 These examples show that C. albicans specific Th17 cells are able to exhibit extensive effector functions towards numerous commensal microorganisms, both in health and in disease.
The effect of fungi-specific Th17 cells on inflammatory diseases is not one-dimensional and likely depends not only on phenotype, but also on environmental signals and location. Traditionally, T helper cells have been divided into pathogenic and non-pathogenic phenotypes, a view which is largely based on in vitro experiments and disregards the high plasticity exhibited by T helper cells, especially in Tregs and Th17 cells, which are of particular importance for antifungal immunity as described above. While C. albicans specific Th17 cells classically have been described exhibiting a pathogenic phenotype,85 they do not cause intestinal inflammation in humans under normal circumstances. More so, while Th17 responses are increased in IBD patients70 and their frequency correlates with the fecal number of C. albicans in intensive care unit patients,86 an IL-17 blockade led to a negative effect for IBD patients. This suggests that Th17 responses against C. albicans are at least partially protective in the inflamed gut, rather than pathogenic. Under homeostatic conditions, IL-17 production in response to commensal fungi may regulate epithelial barrier integrity and/or antimicrobial peptide (AMP) production while strengthening barrier repair functions to promote microbial homeostasis including the equilibrium between bacterial and fungal commensals.
Pathogenic functions of commensal-specific Th17 cells usually arise with altered environmental signals. In mice, recent evidence suggests SAA proteins 1, 2, and 3 have a central role in alternating development of naive T cells towards a pathogenic phenotype characterized by T-bet activity in combination with RORγt.87 In contrast, the observation that local SAA 1 and 2 in the intestine strengthen the barrier function by upregulating IL-1787 highlights not only the importance of environmental signaling, but also the impact of developmental stage of T cells. SAAs thereby could promote a proinflammatory state in naive T cells, but mediate gut homeostasis for memory T cells. Other influencing factors leading to pathogenic function might be T cells’ location within the body and the type of target. C. albicans specific Th17 cross-reacting to Aspergillus fumigatus have been found to contribute to the inflammatory state in the human lung during aspergillosis (Figure 2).70 Also, cross-reactivity to autoantigens82 and allergens88 has been reported in humans, so it might be interesting to further investigate the role of fungal specific Th17 cells in the corresponding diseases.
Viruses
Commensal microbiota have been found to form close relationships with invading viruses. In recent years, it has been proposed that these interactions might dictate the severity and outcome of an infection to a significant degree.1 Additionally, the presence of viruses in healthy humans has been reported, creating intrigue in their role as a commensal.89,90
Yang et al states that gut resident bacteriophages protect from intestinal inflammation.91 An increase in IFN-γ producing CD4+ and CD8+ T cells was found in the Peyer’s patches of GF mice treated with specific purified E. coli phages.92 The group further displayed that DCs incubated with phages stimulated CD4+ T cell production of IFN-γ, suggesting an induction of Th1 cells by bacteriophages.
While it seems reasonable that intestinal phage populations influence gut immunity indirectly by interfering with the growth of their microbial targets, promoting the expansion of other microorganisms or resistant strains, Liu et al 2019 have shown a more direct interaction in mice—commensal viruses maintaining IELs and intestinal immunity, an effect that seems to be of significant importance.93 In addition, potential interactions of commensal viruses with classical CD8+ cytotoxic T cells might also play a role in maintaining homeostasis. Commensal viruses and their interaction with the human immune system thereby pose an interesting and challenging field of study, as both the indirect influence via their bacterial hosts as well as the direct mechanisms, such as those mentioned above, must be considered to fully understand the role of viruses in intestinal immunity. Major challenges arise not only from the need to differentiate between these two effects, but also from the necessity to establish which virus strains are considered commensal and the attached sequencing strategies. More in vitro and in vivo studies on how commensal viruses, such as bacteriophages, can directly and indirectly affect immune cells are needed since data in this field is still limited when compared to other classes of microbes. The key microbes and the induction of CSTCs are summarized in Figure 3.
CSTCs and Disease
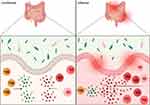
IBD
Given the very high presence of microbes in the intestine, a highly sensitive system of regulation is needed to prevent inflammation. Per the previous sections, the microbial interactions present in the gut are known to aid in maintaining homeostasis; however, the mechanisms through which this is achieved, and where it goes wrong leading to disease, are still not fully elucidated. Many believe CSTC responses may play a major homeostatic role and thus also play a role in disease. CSTC’s role in homeostasis could take the form of a Treg phenotype, for example.65,94 Data suggests the existence of bacterial-specific regulatory T cells and hints at their role in inhibiting the development of colitis in mice.95,96 Commensal bacteria have been found to induce a unique RORγt+ Treg population that specifically induced a homeostatic immune response in mice.96 As mentioned above, previous studies have shown that Clostridia and Schadler’s flora stimulate colonic Tregs.40,56 A Treg-IgA axis specific for flagellin has also been proposed as a key player in homeostatic control of CD4+ T cell responses in the mouse intestine.65 Furthermore, c-MAF, an important transcription factor for Th17 cells, was found to be an important driver of Treg and Th17 cell homeostasis when studying Helicobacter hepaticus specific inflammation in mice.94
Regarding disease, it seems that the mutualistic relationship between commensals and the intestinal immune system could be playing a major role in the pathogenicity observed in Crohn’s disease and ulcerative colitis, together known as inflammatory bowel disease (IBD).97 IBD is characterized by chronic intestinal inflammation. CSTCs have been proposed to be part of the immune dysregulation theory of IBD and are increasingly being considered as key players in IBD development.2,97 C3H/HeJBir is a strain of mice that is highly susceptible to spontaneous development of colitis.98 The CD4+ T cells of these mice seem to be mainly reactive to enteric bacterial flora antigens.23 Furthermore, the same group found that transferring CD4+ T cell lines specific for commensal enteric bacteria of C3H/HeJBir mice to severe combined immunodeficiency (SCID) mice induced colitis.99 There is also evidence that bacterial flagellin could be a major Crohn’s disease antigen and flagellin-specific CD4+ T cells were found to induce severe colitis when also transferred to SCID mice.100 Recent data is also hinting at a major role of RORγt influence on Th17 cells and their connection to inflammation. To maintain pathogenicity of Th17 cells, RORγt was found to be inhibiting IL-10 production, and RORγt inhibition in Th17 cells was connected with less severe colitis in mice.101
Specific, context-dependent mechanisms may be important factors in IBD development and overall inflammation is brought about by an imbalance of CSTCs.4,96 While there are different aspects of the intestinal immune system that have been found to play an important role in immune homeostasis of the gut, and development of inflammation, the overall mechanisms are still unknown. However, CSTCs’ imbalance may be playing a major role (Figure 4). Whether their actions during inflammation are the cause or effect of inflammation, however, has yet to be fully understood.
Autoimmune Diseases
Apart from IBD, CSTCs could also play a role in autoimmune diseases, conditions that often involve a change in the balance between Th cell subtypes and Tregs leading to inflammation. For example, patients with elevated multiple sclerosis (MS) activity, a devastating autoimmune disease in which the myelin of nerve fibers is destroyed by the immune system, are found to have increased Th17 cells in the intestine as well as in the central nervous system with an additional increase being observed during phases of high disease activity.102,103 This effect correlates with an imbalance of intestinal microbiota including a higher Firmicutes/Bacteroidetes ratio, increased relative abundance of Streptococcus, and decreased Prevotella strains compared to healthy controls and MS patients with no disease activity.103 In other studies, an increase in Akkermansia muciniphila and Acinetobacter calcoaceticus, was associated with MS in patients, both pathogens were able to induce proinflammatory responses in human peripheral blood mononuclear cells (PBMCs) as well as in mono-colonized mice.104 Conversely, Parabacteroides distasonis, shown to be decreased in MS patients, has potential protective functions by stimulating anti-inflammatory IL10 expressing human T cells and Tregs in mice.104 While these effects might depend mostly on TCR independent mechanisms,105 it also has been shown that certain viral and bacterial peptides are capable of effectively activating myelin basic protein-specific T cell clones from MS patients via molecular mimicry.106 This suggests a barrier site TCR-dependent expansion of T cells with autoinflammatory effects elsewhere in the body, such as the central nervous system. It might therefore be possible that CSTCs have a significant impact on the development of autoimmune diseases in the central nervous system, such as MS.
Similar to MS, patients with rheumatoid arthritis (RA) also have been found to have increased proportions of Th17 cells and decreased proportions of Tregs in their blood.107 Similar to findings observed by Cekanaviciute et al in MS, the abundance of Prevotella in the intestine has been linked to RA, suggesting potential mechanisms of CSTC activation in the gut leading to autoimmunity. However, while recognizing the importance of the microbiota for the development of RA, TLR activation108,109 has been suggested as an alternative explanation by other studies. Additional studies are needed to further elucidate the role of CSTC-independent vs commensal-specific autoimmune effects that Th17 cells, Tregs, and microbiota exhibit during the development of autoimmune diseases such as MS and RA.
Antiphospholipid syndrome (APS) occurs when the immune system creates antibodies that increase the likelihood of clotting. It has been shown that patients with APS who had immune recognition of gut commensal bacteria, R. intestinalis, had intestinal inflammation sub clinically.110 Th1 cell clones specific to an autoantigen of APS cross-reacted with R. intestinalis mimotopes, driving chronic autoimmunity in APS patients.110 Ruff et al acknowledge that a complex relationship in humans exists between antigen-specific responses, genetic predisposition, and recurring innate-immune driven events against chronically colonizing commensals, which drives repetitive tissue damage from autoreactive B and T cells.110 Chronic autoimmune diseases will be better understood with more studies uncovering the molecular mechanisms between the host immune system and microbiota.
Conclusion and Future Directions
The impact of CSTCs is critical regarding the trillions of microorganisms that colonize the human intestinal tract. In this review, the generation of CSTCs and their various subtypes was examined to better understand their roles in modulating immune development and immune responses. CSTCs exist as CD4+ or CD8+ T cells and contribute to homeostasis at barrier sites such as the intestine to prevent pathogenic inflammatory responses. Numerous organisms within the human and mouse microbiotas play key roles in this commensal relationship, including bacteria, fungi, and viruses. Without the immunological balance provided by certain CSTCs, there is an increase in pathologies leading to intestinal inflammation, such as Crohn’s disease and ulcerative colitis. Moreover, circumstantial evidence suggests that CSTCs can potentially contribute to autoimmune disease such as MS and RA. Collectively, these findings provide further support for the potential impact of CSTCs in maintaining healthy immune responses. More research is required on CSTCs and their impact on intestinal disease, as well as the potential relationship between commensals and other types of immune cells in contributing to inflammation in these contexts. Future studies should consider the influence of microbiota on different T helper cell subsets to study their impact on the development of different inflammatory conditions, including IBD and autoimmune diseases.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
L.K. is supported by start-up funds from Yale University, Binational Science Foundation award number 2019075, and National Institute of Health NIH/NCATS R21TR002639, NIH/NICHD R21HD102565, and NIH/NIAID R01AI171980. A.O. is supported by NIH/NDDK 1K08DK120871-01A1.
Disclosure
Arne Gehlhaar, Ashwin Inala, Dhana Llivichuzhca-Loja, and Tatiana N. Silva are co-first authors for this study. Amy E. O’Connell and Liza Konnikova are co-senior authors for this study. The authors have declared that no conflict of interest exists in this work.
References
1. Li N, Ma W-T, Pang M, Fan Q-L, Hua J-L. The commensal microbiota and viral infection: a comprehensive review. Front Immunol. 2019;10:1551. doi:10.3389/fimmu.2019.01551
2. Sorini C, Cardoso RF, Gagliani N, Villablanca EJ. Commensal bacteria-specific CD4+ T cell responses in health and disease. Front Immunol. 2018;9:2667. doi:10.3389/fimmu.2018.02667
3. Harrison OJ, Linehan JL, Shih H-Y, et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 2019;363:eaat6280. doi:10.1126/science.aat6280
4. Hegazy AN, West NR, Stubbington MJT, et al. Circulating and tissue-resident CD4+ T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. 2017;153:1320–1337.e16. doi:10.1053/j.gastro.2017.07.047
5. Diehl GE, Longman RS, Zhang J-X, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi:10.1038/nature11809
6. Goto Y. Epithelial cells as a transmitter of signals from commensal bacteria and host immune cells. Front Immunol. 2019;10:2057. doi:10.3389/fimmu.2019.02057
7. Park J-H, Kotani T, Konno T, et al. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS One. 2016;11:e0156334. doi:10.1371/journal.pone.0156334
8. Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. The who’s who of T-cell differentiation: human memory T-cell subsets. Eur J Immunol. 2013;43:2797–2809. doi:10.1002/eji.201343751
9. Belkaid Y, Bouladoux N, Hand TW. Effector and memory T cell responses to commensal bacteria. Trends Immunol. 2013;34:299–306. doi:10.1016/j.it.2013.03.003
10. Zegarra-Ruiz DF, Kim DV, Norwood K, et al. Thymic development of gut-microbiota-specific T cells. Nature. 2021;594:413–417. doi:10.1038/s41586-021-03531-1
11. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol. 2014;14:377–391. doi:10.1038/nri3667
12. Pathak M, Lal G. The regulatory function of CCR9+ dendritic cells in inflammation and autoimmunity. Front Immunol. 2020;11:536326. doi:10.3389/fimmu.2020.536326
13. Fujimoto K, Karuppuchamy T, Takemura N, et al. A new subset of CD103 + CD8α + dendritic cells in the small intestine expresses TLR3, TLR7, and TLR9 and induces Th1 response and CTL activity. J Immunol. 2011;186:6287–6295. doi:10.4049/jimmunol.1004036
14. Kayama H, Takeda K. Functions of innate immune cells and commensal bacteria in gut homeostasis. J Biochem. 2016;159:141–149. doi:10.1093/jb/mvv119
15. Birchenough GMH, Nyström EEL, Johansson MEV, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–1542. doi:10.1126/science.aaf7419
16. Sicard J-F, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol. 2017;7:387. doi:10.3389/fcimb.2017.00387
17. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi:10.1016/j.cell.2004.07.002
18. Lei-Leston AC, Murphy AG, Maloy KJ. Epithelial cell inflammasomes in intestinal immunity and inflammation. Front Immunol. 2017;8:1168. doi:10.3389/fimmu.2017.01168
19. Wlodarska M, Thaiss C, Nowarski R, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi:10.1016/j.cell.2014.01.026
20. Zong X, Fu J, Xu B, Wang Y, Jin M. Interplay between gut microbiota and antimicrobial peptides. Anim Nutr. 2020;6:389–396. doi:10.1016/j.aninu.2020.09.002
21. Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol. 2012;3:310. doi:10.3389/fimmu.2012.00310
22. Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi:10.1016/j.immuni.2009.08.020
23. Cong Y, Brandwein SL, McCabe RP, et al. CD4+ T cells reactive to enteric bacterial antigens in spontaneously colitic C3H/HeJBir mice: increased T helper cell type 1 response and ability to transfer disease. J Exp Med. 1998;187:855–864. doi:10.1084/jem.187.6.855
24. Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. 2010;207:1321–1332. doi:10.1084/jem.20092253
25. Imam T, Park S, Kaplan MH, Olson MR. Effector T helper cell subsets in inflammatory bowel diseases. Front Immunol. 2018;9:1212. doi:10.3389/fimmu.2018.01212
26. Walker JA, McKenzie ANJ. TH2 cell development and function. Nat Rev Immunol. 2018;18:121–133. doi:10.1038/nri.2017.118
27. Lundell A-C, Andersson K, Josefsson E, Steinkasserer A, Rudin A. Soluble CD14 and CD83 from human neonatal antigen-presenting cells are inducible by commensal bacteria and suppress allergen-induced human neonatal Th2 differentiation. Infect Immun. 2007;75:4097–4104. doi:10.1128/IAI.01744-06
28. Wu W, Liu H-P, Chen F, et al. Commensal A4 bacteria inhibit intestinal Th2-cell responses through induction of dendritic cell TGF-β production. Eur J Immunol. 2016;46:1162–1167. doi:10.1002/eji.201546160
29. Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi:10.1016/j.cell.2015.08.058
30. Sano T, Huang W, Hall J, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163:381–393. doi:10.1016/j.cell.2015.08.061
31. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi:10.1016/j.cell.2009.09.033
32. Lécuyer E, Rakotobe S, Lengliné-Garnier H, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi:10.1016/j.immuni.2014.03.009
33. Hepworth MR, Fung TC, Masur SH, et al. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria–specific CD4 + T cells. Science. 2015;348:1031–1035. doi:10.1126/science.aaa4812
34. Eyerich K, Foerster S, Rombold S, et al. Patients with chronic mucocutaneous candidiasis exhibit reduced production of Th17-associated cytokines IL-17 and IL-22. J Invest Dermatol. 2008;128:2640–2645. doi:10.1038/jid.2008.139
35. Li J, Casanova J-L, Puel A. Mucocutaneous IL-17 immunity in mice and humans: host defense vs. excessive inflammation. Mucosal Immunol. 2018;11:581–589. doi:10.1038/mi.2017.97
36. Liang SC, Tan X-Y, Luxenberg DP, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi:10.1084/jem.20061308
37. Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi:10.1126/science.1195568
38. Ai TL, Solomon BD, Hsieh C-S. T-cell selection and intestinal homeostasis. Immunol Rev. 2014;259:60–74. doi:10.1111/imr.12171
39. Nutsch K, Chai J, Ai T, et al. Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Rep. 2016;17:206–220. doi:10.1016/j.celrep.2016.08.092
40. Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science. 2011;331:337–341. doi:10.1126/science.1198469
41. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. doi:10.1111/imr.12567
42. Russler-Germain EV, Rengarajan S, Hsieh C-S. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 2017;10:1375–1386. doi:10.1038/mi.2017.65
43. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi:10.1073/pnas.0909122107
44. Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi:10.1038/nature10434
45. Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi:10.1146/annurev.immunol.25.022106.141623
46. Chen B, Ni X, Sun R, et al. Commensal bacteria-dependent CD8αβ+ T cells in the intestinal epithelium produce antimicrobial peptides. Front Immunol. 2018;9:1065. doi:10.3389/fimmu.2018.01065
47. Sankaran-Walters S, Hart R, Dills C. Guardians of the gut: enteric defensins. Front Microbiol. 2017;8:647. doi:10.3389/fmicb.2017.00647
48. Tanoue T, Morita S, Plichta DR, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600–605. doi:10.1038/s41586-019-0878-z
49. Yu AI, Zhao L, Eaton KA, et al. Gut microbiota modulate CD8 T cell responses to influence colitis-associated tumorigenesis. Cell Rep. 2020;31:107471. doi:10.1016/j.celrep.2020.03.035
50. Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi:10.1038/nri3007
51. O’Callaghan J, O’Toole PW. Lactobacillus: host-microbe relationships. Curr Top Microbiol Immunol. 2013;358:119–154. doi:10.1007/82_2011_187
52. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108:4680–4687. doi:10.1073/pnas.1002611107
53. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in Newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi:10.1073/pnas.1002601107
54. Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi:10.1186/1757-4749-5-23
55. Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K, McGhee JR. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–3511. doi:10.1128/IAI.67.7.3504-3511.1999
56. Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi:10.1038/nature12331
57. Watanabe M, Ueno Y, Yajima T, et al. Interleukin 7 is produced by human intestinal epithelial cells and regulates the proliferation of intestinal mucosal lymphocytes. J Clin Invest. 1995;95:2945–2953. doi:10.1172/JCI118002
58. Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–217. doi:10.1038/nrmicro2298
59. Benno Y, Sawada K, Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28:975–986. doi:10.1111/j.1348-0421.1984.tb00754.x
60. Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15:189–203. doi:10.1099/00222615-15-2-189
61. Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–321. doi:10.1542/peds.72.3.317
62. Erdogan A, Rao SSC. Small intestinal fungal overgrowth. Curr Gastroenterol Rep. 2015;17:16. doi:10.1007/s11894-015-0436-2
63. Kumamoto CA. Candida biofilms. Curr Opin Microbiol. 2002;5:608–611. doi:10.1016/S1369-5274(02)00371-5
64. Kondori N, Nowrouzian F, Ajdari M, et al. Candida species as commensal gut colonizers: a study of 133 longitudinally followed Swedish infants. Med Mycol. 2020;58:485–492. doi:10.1093/mmy/myz091
65. Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci. 2009;106:19256–19261. doi:10.1073/pnas.0812681106
66. Uchida AM, Boden EK, James EA, et al. Escherichiacoli-specific CD4+ T cells have public T-cell receptors and low interleukin 10 production in Crohn’s disease. Cell Mol Gastroenterol Hepatol. 2020;10:507–526. doi:10.1016/j.jcmgh.2020.04.013
67. Kapitan M, Niemiec MJ, Steimle A, Frick JS, Jacobsen ID. Fungi as part of the microbiota and interactions with intestinal bacteria. Curr Top Microbiol Immunol. 2019;422:265–301. doi:10.1007/82_2018_117
68. Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–358. doi:10.1080/21505594.2016.1247140
69. Nash AK, Auchtung TA, Wong MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi:10.1186/s40168-017-0373-4
70. Bacher P, Hohnstein T, Beerbaum E, et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell. 2019;176:1340–1355.e15. doi:10.1016/j.cell.2019.01.041
71. Blaschke-Hellmessen R. [Vertical transmission of Candida and its consequences]. Mycoses. 1998;41(Suppl 2):31–36. doi:10.1111/j.1439-0507.1998.tb00598.x
72. Jo J-H, Deming C, Kennedy EA, et al. Diverse human skin fungal communities in children converge in adulthood. J Investig Dermatol. 2016;136:2356–2363. doi:10.1016/j.jid.2016.05.130
73. Vogel K, Pierau M, Arra A, et al. Scientific reports. Developmental induction of human T-cell responses against Candida albicans and Aspergillus fumigatus. Available from: https://www.nature.com/articles/s41598-018-35161-5.
74. Acosta-Rodriguez EV, Rivino L, Geginat J, et al. Surface phenotype and antigenic specificity of human interleukin 17–producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi:10.1038/ni1467
75. Bacher P, Schink C, Teutschbein J, et al. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J Immunol. 2013;190:3967–3976. doi:10.4049/jimmunol.1202221
76. Bacher P, Kniemeyer O, Schönbrunn A, et al. Antigen-specific expansion of human regulatory T cells as a major tolerance mechanism against mucosal fungi. Mucosal Immunol. 2014;7:916–928. doi:10.1038/mi.2013.107
77. Elsevier Enhanced Reader. T cell immunity to commensal fungi. Available from: https://reader.elsevier.com/reader/sd/pii/S1369527420301120?token=8725BB8F03A05C31C2119B026E9247696888C16F657B37B62AE6AE795BA0CA7BC271B9E910C33D9D2C85B4E5DF4EC492&originRegion=us-east-1&originCreation=20211019153526.
78. Elsevier Enhanced Reader. CD4+CD25+Foxp3+ regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Available from: https://reader.elsevier.com/reader/sd/pii/S107476131100080X?token=B1A2DE9A72EA5BD418D32344CABFB1C1C17AFBCAC9AD9841CA3809A1FACB5673377598F8BBC105BD38B61DDCD2C2F849&originRegion=us-east-1&originCreation=20211019214506.
79. Kirchner FR, Littringer K, Altmeier S, et al. Persistence of Candida albicans in the oral mucosa induces a curbed inflammatory host response that is independent of immunosuppression. Front Immunol. 2019;10. doi:10.3389/fimmu.2019.00330
80. Kurup VP, Seymour BW, Choi H, Coffman RL. Particulate Aspergillus fumigatus antigens elicit a TH2 response in BALB/c mice. J Allergy Clin Immunol. 1994;93:1013–1020. doi:10.1016/S0091-6749(94)70050-8
81. Huang X, Johansson SG, Zargari A, Nordvall SL. Allergen cross-reactivity between Pityrosporum orbiculare and Candida albicans. Allergy. 1995;50:648–656. doi:10.1111/j.1398-9995.1995.tb02581.x
82. Ruff WE, Greiling TM, Kriegel MA. Host-microbiota interactions in immune-mediated diseases. Nat Rev Microbiol. 2020;18:521–538. doi:10.1038/s41579-020-0367-2
83. Koivikko A, Kalimo K, Nieminen E, et al. Allergenic cross-reactivity of yeasts. Allergy. 1988;43:192–200. doi:10.1111/j.1398-9995.1988.tb00418.x
84. Zielinski CE, Mele F, Aschenbrenner D, et al. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484:514–518. doi:10.1038/nature10957
85. Noster R, de Koning HD, Maier E, et al. Dysregulation of proinflammatory versus anti-inflammatory human TH17 cell functionalities in the autoinflammatory Schnitzler syndrome. J Allergy Clin Immunol. 2016;138:1161–1169.e6. doi:10.1016/j.jaci.2015.12.1338
86. Shao T-Y, Ang WXG, Jiang TT, et al. Commensal Candida albicans positively calibrates systemic Th17 immunological responses. Cell Host Microbe. 2019;25:404–417.e6. doi:10.1016/j.chom.2019.02.004
87. Lee J-Y, Hall JA, Kroehling L, et al. Serum amyloid A proteins induce pathogenic TH17 cells and promote inflammatory disease. Cell. 2020;180:79–91.e16. doi:10.1016/j.cell.2019.11.026
88. Su LF, Davis MM. Antiviral memory phenotype T cells in unexposed adults. Immunol Rev. 2013;255:95–109. doi:10.1111/imr.12095
89. Sauvage V, Cheval J, Foulongne V, et al. Identification of the first human gyrovirus, a virus related to chicken anemia virus. J Virol. 2011;85:7948–7950. doi:10.1128/JVI.00639-11
90. Kapusinszky B, Minor P, Delwart E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. 2012;50:3427–3434. doi:10.1128/JCM.01589-12
91. Yang J-Y, Kim M-S, Kim E, et al. Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-β production. Immunity. 2016;44:889–900. doi:10.1016/j.immuni.2016.03.009
92. Gogokhia L, Buhrke K, Bell R, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019;25:285–299.e8. doi:10.1016/j.chom.2019.01.008
93. Liu L, Gong T, Tao W, et al. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat Immunol. 2019;20:1681–1691. doi:10.1038/s41590-019-0513-z
94. Xu M, Pokrovskii M, Ding Y, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018;554:373–377. doi:10.1038/nature25500
95. Cong Y, Weaver CT, Lazenby A, Elson CO. Bacterial-reactive T regulatory cells inhibit pathogenic immune responses to the enteric flora. J Immunol. 2002;169:6112–6119. doi:10.4049/jimmunol.169.11.6112
96. Sefik E, Geva-Zatorsky N, Oh S, et al. Individual intestinal symbionts induce a distinct population of RORγ + regulatory T cells. Science. 2015;349:993–997. doi:10.1126/science.aaa9420
97. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi:10.1146/annurev-immunol-030409-101225
98. Elson CO, Cong Y, Sundberg J. The C3H/HeJBir mouse model: a high susceptibility phenotype for colitis. Int Rev Immunol. 2000;19:63–75. doi:10.3109/08830180009048390
99. Cong Y, Weaver CT, Lazenby A, Elson CO. Colitis induced by enteric bacterial antigen-specific CD4+ T cells requires CD40-CD40 ligand interactions for a sustained increase in mucosal IL-12. J Immunol. 2000;165:2173–2182. doi:10.4049/jimmunol.165.4.2173
100. Lodes MJ, Cong Y, Elson CO, et al. Bacterial flagellin is a dominant antigen in Crohn disease; 2004. Available from: https://www.jci.org/articles/view/20295/pdf.
101. Sun M, He C, Chen L, et al. RORγt represses IL-10 production in Th17 cells to maintain their pathogenicity in inducing intestinal inflammation. J Immunol 2019;202:79–92.
102. Moser T, Akgün K, Proschmann U, Sellner J, Ziemssen T. The role of TH17 cells in multiple sclerosis: therapeutic implications. Autoimmun Rev. 2020;19:102647. doi:10.1016/j.autrev.2020.102647
103. Cosorich I, Dalla-Costa G, Sorini C, et al. High frequency of intestinal T H 17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. 2017;3:e1700492. doi:10.1126/sciadv.1700492
104. Cekanaviciute E, Yoo BB, Runia TF, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114:10713–10718. doi:10.1073/pnas.1711235114
105. Haase S, Haghikia A, Wilck N, Müller DN, Linker RA. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology. 2018;154:230–238. doi:10.1111/imm.12933
106. Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi:10.1016/0092-8674(95)90348-8
107. Samson M, Audia S, Janikashvili N, et al. Brief report: inhibition of interleukin-6 function corrects Th17/Treg cell imbalance in patients with rheumatoid arthritis. Arthritis Rheum. 2012;64:2499–2503. doi:10.1002/art.34477
108. Rogier R, Koenders MI, Abdollahi-Roodsaz S. Toll-like receptor mediated modulation of T cell response by commensal intestinal microbiota as a trigger for autoimmune arthritis. J Immunol Res. 2015;2015:e527696. doi:10.1155/2015/527696
109. Abdollahi-Roodsaz S, Joosten LAB, Koenders MI, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118:205–216. doi:10.1172/JCI32639
110. Ruff WE, Dehner C, Kim WJ, et al. Pathogenic autoreactive T and B cells cross-react with mimotopes expressed by a common human gut commensal to trigger autoimmunity. Cell Host Microbe. 2019;26:100–113.e8. doi:10.1016/j.chom.2019.05.003
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.