Back to Journals » OncoTargets and Therapy » Volume 12
INPP4B As A Prognostic And Diagnostic Marker Regulates Cell Growth Of Pancreatic Cancer Via Activating AKT
Authors Zhai S , Liu Y, Lu X, Qian H, Tang X, Cheng X, Wang Y, Shi Y, Deng X
Received 15 July 2019
Accepted for publication 24 September 2019
Published 9 October 2019 Volume 2019:12 Pages 8287—8299
DOI https://doi.org/10.2147/OTT.S223221
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Shuyu Zhai,* Yuanbin Liu,* Xiongxiong Lu,* Hao Qian, Xiaomei Tang, Xi Cheng, Yue Wang, Yusheng Shi, Xiaxing Deng
Department of General Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaxing Deng; Yusheng Shi
Department of General Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, 197 RuiJin 2 Road, Shanghai 200025, People’s Republic of China
Email [email protected]; [email protected]
Background: Inositol polyphosphate 4-phosphatase type II (INPP4B), a member of the PI3K/Akt signaling pathway, plays a vital role in the initiation and progression of cancers. However, its biological role in pancreatic cancer remains largely undiscovered. Our study aimed to investigate the effects of INPP4B on proliferation in pancreatic cancer and its clinical relevance.
Materials and methods: INPP4B expression data were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. Clinicopathological and survival data were retrieved from the TCGA database. CCK8 and colony formation assays were performed to measure the proliferative capacity of pancreatic cancer. Tumor xenograft models were established to measure cancer proliferative abilities in vivo.
Results: INPP4B was upregulated in pancreatic cancer tissue compared with normal tissue. INPP4B knockdown inhibited cell proliferation and promoted apoptosis in pancreatic cancer in vitro and in vivo. INPP4B knockdown also reduced AKT phosphorylation. Moreover, INPP4B was associated with poor overall and disease-free survival, with Cox regression analysis showing that INPP4B could serve as an independent prognostic marker. ROC curve analysis showed that INPP4B possessed moderate diagnostic value.
Conclusion: Collectively, INPP4B is an oncogenic gene in pancreatic cancer and could serve as a potential diagnostic marker and an independent prognostic marker, suggesting that it could be a novel therapeutic target for pancreatic cancer.
Keywords: INPP4B, pancreatic cancer, proliferation, prognosis, diagnosis
Introduction
Pancreatic cancer is one of the most lethal tumors characterized by late diagnosis and poor prognosis, with a 5-year survival less than 5%.1 Chemotherapy is not always effective for patients with pancreatic cancer due to profound drug resistance.2–4 Therefore, it is imperative to discover novel targets to increase the early diagnostic rate and improve the prognosis of pancreatic cancer patients.
Precision medicine and target therapy are currently gaining more interest.5 Extensive research has been focused on the identification of clinically useful markers for pancreatic cancer. In addition to CA19-9 and CA125, which have been well-demonstrated as diagnostic and prognostic markers for pancreatic cancer,6,7 MIC-1, PAM4, S100A6 and OPN are newly discovered diagnostic markers, while SPARC, CISD2 and SMAD4 have been identified as promising prognostic markers.8–14
INPP4B, inositol polyphosphate 4‐phosphatase type II, is a phosphoinositide phosphatase that hydrolyzes PI(3,4,5)P2 to PI(3,4)P and can interact with the AKT pleckstrin homology domain to induce full activation of AKT.15 Previous studies have proven that INPP4B expression was inhibited in thyroid cancer, melanoma, prostate cancer and breast cancer. Reduced INPP4B could enhance cell proliferation and migration by deactivating AKT, suggesting its tumor suppressor role in these cancer cells.16–19 However, some research has oppositely reported that INPP4B could serve as an oncogenic factor in colon cancer, acute myeloid leukemia and in a subset of melanomas.20–22 Briefly, the role of INPP4B in carcinogenesis and cancer progression still remains controversial. Recent evidence has demonstrated that increased INPP4B expression is associated with poor clinical outcomes in pancreatic cancer.23 Interestingly, another study has suggested that INPP4B could inhibit the epithelial-mesenchymal transition and invasive capacity of pancreatic cancer via inactivating AKT.24 In our study, however, we indicated an opposite role of INPP4B in pancreatic cancer.
In this study, we downloaded and analyzed expression and clinicopathological data from the TCGA and GEO databases. Combined with biological validation, we proved that INPP4B was significantly upregulated in pancreatic cancer. INPP4B knockdown inhibited cell proliferation and promoted apoptosis of pancreatic cancer both in vitro and in vivo. Moreover, INPP4B knockdown inhibited AKT phosphorylation. This was the first time that we reported that INPP4B possessed a moderate diagnostic value and that INPP4B overexpression was associated with poor patient survival, suggesting that INPP4B could be an oncogene and potentially serve as a diagnostic and prognostic marker for pancreatic cancer.
Materials And Methods
Bioinformatics Analysis
Expression And Survival Data Extraction
INPP4B expression data from pancreatic cancer and normal tissues were obtained and analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) server (http://gepia.cancer-pku.cn/index.html),25 which is based on the Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) project, including 179 cancer and 171 normal samples. We also downloaded and extracted the expression data from four GEO datasets (GSE16515, GSE15471, GSE28735 and GSE62452) that compare mRNA expression between normal and pancreatic cancer tissues; three GEO datasets (GSE101462, GSE91035 and GSE77858) that compare the expression among normal, pancreatitis and pancreatic cancer tissues; and one dataset from Oncomine uploaded by Logsdon. GSE16515 contains 36 pancreatic cancer samples and 16 normal samples. GSE15471 contains 36 cancer tissues and 36 normal tissues. GSE28735 includes 45 cancer tissues and 45 normal tissues. GSE62452 includes 69 cancer samples and 61 normal samples. GSE101462 contains 4 normal tissues, 10 pancreatitis tissues and 6 tumor tissues. GSE91035 includes 8 normal samples, 17 pancreatitis and benign pancreatic tissues, and 25 tumor samples. GSE77858 contains 3 normal tissues, 5 pancreatitis tissues and 77 tumor tissues. The dataset from Oncomine includes 5 normal and pancreatitis tissues, and 17 tumor tissues. Clinicopathological and survival data of 183 patients from the TCGA datasets were downloaded and analyzed. Excluding patients with incomplete INPP4B expression data or clinicopathological data, a total of 176 patients were included to perform survival analysis and divided into two groups (88 patients with high INPP4B expression levels and 88 with low expression levels); moreover, a total of 162 patients were included in the clinicopathological correlation analysis. To determine the diagnostic value, we analyzed two other GEO datasets (GSE1542 and GSE71729). GSE1542 contains 24 pancreatic cancer samples and 25 normal samples, while GSE71729 includes 145 cancer tissues and 46 normal tissues.
KEGG Pathway Enrichment Analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database that contains various kinds of data from large molecular datasets generated using high-throughput experimental technologies.26 The top 100 coexpressed genes targeted by INPP4B in pancreatic cancer were screened using the cBioPortal (http://www.cbioportal.org).27 KEGG pathway enrichment analysis of these 100 genes was performed using the clusterProfiler package in R (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html).28 The results were visualized using GraphPad Prism 7.
Clinical Samples
Written informed consents were obtained from all the participating patients and the experiments involving in human specimens were conducted in compliance with the Ethics Committee of Human Experimentation and with the Declaration of Helsinki as revised in 2013. The study protocol was approved by the Ethics Committee of Shanghai Ruijin Hospital (No. 121 in 2017) for scientific research on these clinical materials. Twenty-two surgical specimens (eleven tumor tissues and paired adjacent tumor tissues) were obtained from patients treated at Ruijin Hospital. The tumor samples used in this study were all derived from pancreatic ductal adenocarcinomas that were independently identified by two expert pathologists.
Cell Culture
Six cell lines derived from pancreatic tumors (Bxpc3, Sw1990, Panc-1, Patu8988 and Capan1) and the human pancreatic ductal epithelial (HPDE) cell line were purchased from the Cell bank of the Chinese Academy of Sciences and were cultured in RPMI 1640, DMEM, and IMDM supplemented with 10% fetal bovine serum and antibiotics.
Quantitative Real-Time PCR
Total RNAs from the cultured cell lines were extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcription reactions were performed using random primers and an M-MLV Reverse Transcriptase kit (Invitrogen). Quantitative real-time PCR was then performed to amplify the cDNAs with the SYBR Green PCR kit (Toyobo, Osaka, Japan) on a Rotor-Gene RG-3000A (Corbett Life Science, Sydney, NSW, Australia). INPP4B expression levels were quantified using the 2−ΔΔCt method. GAPDH was used as the internal control. The primers for INPP4B were 5′‐GCCGACCACATCACCACAG‐3′ (forward) and 5′‐TTTCCGCTCACACTTTCCG‐3′ (reverse) and the primers for GAPDH were 5′‐TCAGCCGCATCTTCTTTTGC‐3′ (forward) and 5′‐GCGCCCAATACGACCAAATC‐3′ (reverse).
Western Blotting
The cell samples were lysed using RIPA buffer (Sigma-Aldrich; R0278) supplemented with protease and phosphatase inhibitor cocktails (Sigma-Aldrich; P8340). The protein samples were separated by 7.5% SDS-PAGE gel and then were transferred onto PVDF membranes. Primary antibodies and corresponding secondary antibodies were used to bind the corresponding proteins. Primary antibodies included INPP4B (Rabbit mAb, 14543S, 1:1000 for Western blotting and 1:800 for IHC; CST, USA), GAPDH (Mouse mAb, 60004-1-Ig, 1:5000; Proteintech, USA), cleaved-PARP (Rabbit mAb, 5625T, 1:1000; CST, USA), cleaved-Caspase 3 (Rabbit mAb, 9664T, 1:1000; CST, USA), Bax (Rabbit mAb, 5023T, 1:1000; CST, USA), p-PI3K (Rabbit mAb, 17366S, 1:1000; CST, USA) and p-AKT (Rabbit mAb, 4060T, 1:1000; CST, USA). Protein binding was quantified using the ImageJ software.
Stable Knockdown Of INPP4B
The shRNA sequences for INPP4B was designed by Jiman Gene (Shanghai, China). The lentiviral vectors carrying INPP4B shRNA were added to patu8988 cultured cell lines for 24 h. Subsequently, puromycin (5.0 μg/mL, Sigma) was added to select for resistant colonies for 2–3 weeks. INPP4B expression levels in the selected cells were detected by qRT-PCR assay and/or Western blot assay.
Stable Overexpression Of INPP4B
The full-length human INPP4B cDNA, generated from Jiman Gene (Shanghai, China), was cloned into the pSuper-flag-puro vector. Cancer cells were transfected with 2 μg/mL INPP4B or vector control DNA with Lipofectamine 2000 (Invitrogen, Shanghai, China). Puromycin (5.0 μg/mL, Sigma) was used to select the stable overexpression colonies for further studies.
siRNA Knockdown Of INPP4B
Three siRNA sequences for INPP4B were obtained from Jima Gene. Transfection of INPP4B siRNA was mediated by Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the distributor’s instructions.
Cell Proliferation Assay
Cell counting kit 8 (CCK8, Dojindo, Japan) was used to measure the cell proliferation capacity. Cells were seeded in 96-well plates at approximately 4000 cells per well and cultured in the appropriate medium. The number of viable cells was quantified by measuring the OD450 at each 24 h interval using a microplate reader (Epoch; BioTek, Winooski, VT). Colony formation assay was performed to confirm the proliferative ability of the pancreatic cancer cells. Cells were seeded into six-well plates with 1000 cells/well in 2 mL culture medium, incubated for 14 days, and stained with 0.1% crystal violet solution. The experiments were performed in triplicate independently.
Tumor Xenograft Assay
Nude BALB/c mice (male, 4–6 weeks old) were purchased from the Chinese Academy of Sciences (Shanghai, China) and were maintained in a specific pathogen-free facility. Patu8988 cells carrying vectors stably inhibiting INPP4B and the normal control were subcutaneously injected into nude mice (5×106 cells/site). Tumor volumes were measured every 4 days to 36 days from the first injection using the following formula: tumor volume (mm3) = 1/2 (a×b2), where “a” represents the longest longitudinal diameter and “b” is the longest transverse diameter. After that, the mice were sacrificed and the tumors were excised. The tumor weights were measured before they were sliced and embedded for immunohistochemistry analysis. All animal experimental procedures were performed in compliance with the institutional ethical requirements and were approved by the Shanghai Jiao Tong University School of Medicine Committee for the Use and Care of Animals.
Immunohistochemical Analysis
The samples were fixed in formalin, embedded and cut into 4-μm sections. The sections were incubated with primary antibody against INPP4B (rabbit monoclonal; ab81269, 1:500; Abcam, USA). Negative controls were incubated without primary antibody. After the slides were stained with DAB and counterstained with hematoxylin, INPP4B staining was examined using ImageJ. The specimens with negative or weak (-/+) INPP4B staining intensity were considered as low INPP4B expression, while those with moderate or strong (++/+++) INPP4B staining intensity were regarded as high INPP4B expression.
Statistical Analysis
SPSS version 20.0 (SPSS Inc., Chicago, IL, USA) was used to perform the statistical analysis. The Pearson χ2 test was used to assess INPP4B expression and its association with clinicopathological characteristics in pancreatic cancer patients. The Student’s t test was used to compare the differences between two groups. Overall and disease-free survival were evaluated using Kaplan-Meier curve analysis by the log rank test. Cox regression analysis was used to assess the 95% confidence intervals (CI) and hazard ratio (HR). The ROC curve was plotted to determine the diagnostic value of INPP4B. The area under the ROC curve (AUC) was used to assess the diagnostic power. A graded AUC value of 0.5–0.7, 0.7–0.9 or 0.9–1.0 represented a poor, moderate or high diagnostic value, respectively. A value of P < 0.05 was considered to denote statistical significance.
Results
INPP4B Was Upregulated In Pancreatic Cancer Tissue
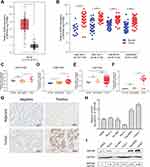
By analyzing the expression data downloaded from the TCGA database and six GEO datasets, we found that INPP4B was significantly upregulated in pancreatic cancer tissue compared with normal tissue (Figure 1A and B). More precisely, through analyzing the expression data of three GEO datasets and one Oncomine dataset, we observed that INPP4B was significantly upregulated in pancreatic cancer tissue compared with pancreatitis or normal tissue (Figure 1C–F). As is shown in Table 1 by immunohistochemistry analysis of 11 pairs of patient samples, we found that moderate or strong staining intensity was found in most pancreatic cancer tissues (8/11), while no or low intensity was detected primarily in normal tissues (9/11), with a significant difference between the two groups (X2=6.6000, P=0.0102). The representative images are displayed in Figure 1G. INPP4B mRNA and protein expression in pancreatic cell lines are shown in Figure 1H and I, respectively.
 |
Table 1 INPP4B Expression In Pancreatic Cancer And Paired Adjacent Tissues By Immunohistochemical Analysis (n=22) |
INPP4B Knockdown Inhibited Cell Proliferation In Pancreatic Cancer
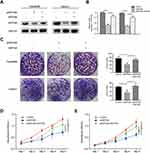
To measure the effect of INPP4B on the proliferation capacity of pancreatic cancer, we divided the patu8988 and capan1 cell lines into three groups that were treated with empty vector, siINPP4B or siINPP4B plus INPP4B plasmid (Figure 2A and B). The colony formation assays showed that INPP4B knockdown could reduce colony formation in patu8988 and capan1 cells, while overexpression of INPP4B could reverse the inhibition of colony formation (Figure 2C). Additionally, CCK8 assay confirmed that reducing INPP4B levels could undermine cell viability in patu8988 and capan1 cells (Figure 2D and E).
INPP4B Knockdown Promoted Apoptosis In Pancreatic Cancer
To investigate the effect of INPP4B on apoptosis in pancreatic cancer, we detected the expression of apoptosis-associated proteins by Western blot. The results showed that the apoptosis-associated proteins cleaved PARP, Caspase-3 and Bax were significantly upregulated in patu8988 cells transfected with siINPP4B (Figure 3A and B).
INPP4B Knockdown Inhibited AKT Phosphorylation
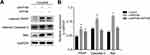
To explore the underlying molecular mechanism through which INPP4B exerts its biological function, KEGG pathway enrichment analysis was performed using the top 100 coexpressed INPP4B genes generated from the cBioPortal. The results were mainly enriched for focal adhesion, cancer pathways and the PI3K-AKT signaling pathway (Figure 4A). To further validate the above results, we detected the expression of phosphorylated PI3K (p-PI3K) and phosphorylated AKT (p-AKT) in patu8988 cells with empty control or siINPP4B. The results showed that p-AKT was significantly reduced in pancreatic cancer cells with siINPP4B, while p-PI3K showed no obvious alterations (Figure 4B). The expression of the above proteins was quantified and is shown in Figure 4C.
AKT Activation Was Necessary For INPP4B-Induced Colony Formation
To investigate the necessity of AKT in INPP4B signal transduction, we compared patu8988 cells with siINPP4B and patu8988 cells with both siINPP4B and the AKT activator SC79. We found that INPP4B knockdown-induced inhibition of colony formation was reversed by AKT activation (Figure 5A). However, the decreased cell viability induced by INPP4B silencing detected by CCK8 was not fully rescued by AKT activation (Figure 5B). Moreover, INPP4B knockdown-induced enhanced apoptosis was also reversed by AKT activation (Figure 5C).
INPP4B Knockdown Inhibited Cell Proliferation And Enhanced Apoptosis In Vivo
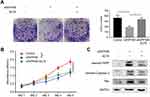
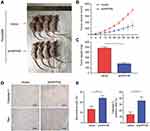
To further validate the vitro analysis results, we established a xenograft model using BALB/c nude mice. Patu8988 cells transfected with vector or shINPP4B were subcutaneously injected into two groups of nude mice. After 36 days, the tumors were presented in Figure 6A. The tumor volumes and weights showed a significant decrease in the shINPP4B group compared with the vector group (Figure 6B and C). Immunohistochemistry analysis of excised tumor tissues showed that the signal intensity of Caspase-3 and Bax in the shINPP4B group was significantly stronger than in the vector group (Figure 6D and E).
INPP4B Could Be A Potential Prognostic And Diagnostic Marker For Pancreatic Cancer
To determine the clinical relevance and prognostic value of INPP4B, we obtained and analyzed clinical data from pancreatic cancer patients from the TCGA database. As is shown in Table 2, high INPP4B expression was correlated with a positive resection margin (R1 or R2, P = 0.008). With respect to its prognostic value, the Kaplan-Meier curve showed that high INPP4B expression in pancreatic cancer patients was associated with poor overall survival (P = 0.0011) and disease-free survival (P = 0.025) (Figure 7A and B). Moreover, as is shown in Table 3, univariate cox regression analysis showed that the T stage (P = 0.049), N stage (P = 0.003), R status (P = 0.009) and INPP4B expression levels (P = 0.003) were closely related to patient survival, while multivariate analysis showed that N stage (P = 0.024) and INPP4B expression levels (P = 0.008) were significantly associated with patient survival, suggesting that INPP4B expression could be an independent prognostic factor for pancreatic cancer. Finally, we evaluated the diagnostic value of INPP4B from the ROC curve using data from six GEO datasets (GSE62452, GSE28735, GSE15471, GSE16515, GSE71729 and GSE1542). The AUC of INPP4B in each dataset was 0.866, 0.878, 0.911, 0.858, 0.779 and 0.642, respectively, which was almost equal to or greater than that of other established diagnostic markers, such as CA125, AFP and CEA (Figure 8A–F), suggesting that INPP4B could be a potential diagnostic marker for pancreatic cancer.
 |
Table 2 The Correlation Between INPP4B Expression And The Clinicopathological Features Of Patients With Pancreatic Cancer |
 |
Table 3 Univariate And Multivariate Analysis Of Clinicopathological Parameters Of Patients With Pancreatic Cancer In The TCGA Database By Cox Regression |
Discussion
Through bioinformatics analysis followed by biological validation, we found that INPP4B was significantly upregulated in pancreatic cancer tissue compared with normal tissue. INPP4B knockdown could reduce the proliferation capacity while promoting the apoptosis of pancreatic cancer cells measured by both in vitro and in vivo analysis. KEGG pathway enrichment analysis using the top 100 coexpressed INPP4B genes showed that the main enriched pathways included the PI3K-AKT signaling pathway. The Western blot results showed that INPP4B knockdown resulted in less phosphorylated AKT, and AKT activation was necessary when INPP4B exerted its oncogenic functions. Furthermore, we identified INPP4B as a potential prognostic and diagnostic marker for pancreatic cancer using data from the TCGA and GEO databases.
In this study, we reported that INPP4B served as an oncogene via AKT activation in pancreatic cancer confirmed by in vitro and in vivo analysis. These results are in contrast to previously published studies suggesting that INPP4B emerged as a tumor suppressor by inactivating AKT in various kind of cancers, such as prostate and basal-like breast cancer.18,19 It has been well-established that INPP4B hydrolyzes PI(3,4)P2, leading to PI3K signaling termination.29 However, the results in our study showed that INPP4B knockdown inhibited and INPP4B overexpression promoted AKT phosphorylation, suggesting that INPP4B promotes AKT activation through upregulation of cellular levels of PI(3,4,5)P3 and PI(3,4)P2. We speculated that the possible mechanism is that while INPP4B hydrolyzes PI(3,4,5)P2 to PI(3,4)P, more PI(3,4,5)P2 molecules are generated through the dephosphorylation of increased PI(3,4)P3 by 5-phosphatases such as PIB5PA and SHIP2.15,30 According to our results, it was confusing that AKT activation could only rescue the inhibition of colony formation induced by INPP4B knockdown, but not the cell viability measured by CCK8, which may result from the inhibition of multiple carcinogenesis signaling pathways by INPP4B silencing, with only the AKT pathway being rescued. However, how AKT activation reversed the inhibition of colony formation induced by INPP4B silencing remains unclear. Therefore, more biological tests should be performed to validate the results using various cell lines.
There are already many molecules that have been identified as reliable diagnostic markers for pancreatic cancer, such as CA19-9, IGF-I and VEGFR2.31 In our study, INPP4B expression in patient samples and cell lines appears to vary widely, perhaps due to the heterogeneous pathophysiological features of patients and pancreatic cancer cell lines that are derived from different cancer locations (primary tumor, liver metastasis or spleen metastasis) in various individuals. In addition, the physiological conditions of cancer cells during cell culture could be affected by many factors such as culture medium or cell generation. Here, we detected the expression of INPP4B in cell lines to select appropriate cell lines with relatively high INPP4B expression level to create knockdown cell models. Importantly, using expression and clinical data from six GEO datasets, we performed ROC analysis to evaluate the diagnostic value of INPP4B. The AUC values were 0.866, 0.878, 0.911, 0.858, 0.779 and 0.642, which were approximately greater than or equal to that of other established markers including CA125, AFP and CEA, suggesting that INPP4B had potential diagnostic value. However, more patient samples are required to validate these results. The identification of novel diagnostic markers may contribute to individualized therapy. We speculate that combination of INPP4B with CA19-9 or another well-established diagnostic marker could improve the sensitivity and specificity of pancreatic cancer detection. However, this requires further investigation to validate with a large patient cohort.
Pancreatic cancer is characterized by its poor prognosis, with a 5-year survival less than 5% for decades. Improving the prognosis of pancreatic cancer patients is difficult for pancreatic surgeons.32 Many prognostic markers in pancreatic cancer have been recognized, such as TRPM8, CD74 and LKB1.33–35 Here, we obtained and analyzed survival data from the GEPIA database, which is based on TCGA and the GTEx project. The results showed that INPP4B was significantly associated with the R status of pancreatic cancer. Additionally, INPP4B overexpression was associated with poor overall and disease-free survival of pancreatic cancer patients, which is in accordance with a previously published research using another analysis tool, suggesting that elevated INPP4B expression was correlated with poor outcomes in pancreatic cancer patients.23 Moreover, multivariate cox regression analysis showed that INPP4B expression and N stage were significantly related to patient survival, indicating that INPP4B could serve as an independent prognostic marker for pancreatic cancer. Taken together, our study suggested that INPP4B could be a potential diagnostic and prognostic marker for pancreatic cancer. However, these results require further studies with a larger patient cohort for validation.
There are some limitations in this study that cannot be neglected. First, the specific molecular mechanisms through which INPP4B promotes rather than inhibits AKT phosphorylation require further elucidation. Second, due to limited number of enrolled patients in the TCGA (176 pancreatic cancer patients) and GEO (patient numbers ranging from 49 to 191) databases, the diagnostic and prognostic potential, as well as the clinical relevance, of INPP4B need to be further evaluated with more patient samples. Moreover, more pancreatitis or benign pancreatic samples are required when comparing with normal and pancreatic cancer samples to detect the specificity of INPP4B as a diagnostic marker. Third, apoptosis in pancreatic cancer was only detected through Western blot by measuring several apoptosis-associated proteins (PARP, Caspase-3 and Bax). Flow cytometry analysis should also be performed to detect the apoptosis rate. Finally, to measure the effect of INPP4B on the proliferation of pancreatic cancer, cell cycle analysis should be performed and the targets that INPP4B regulates in the cell cycle need to be discovered. This is what we plan to investigate in future studies.
Conclusion
Collectively, we proved that INPP4B was upregulated in pancreatic cancer using bioinformatics analysis followed by biological validation. Using in vitro and in vivo analyses, we found that INPP4B could promote proliferation and inhibit apoptosis of pancreatic cancer cells by activating AKT, suggesting that INPP4B is an oncogene in pancreatic cancer. Moreover, INPP4B could serve as an independent prognostic factor and possessed moderate diagnostic value.
Funding
This study was supported by a grant from the Shanghai Anti-cancer Association (EYAS PROJECT; no. SACA-CY1C19).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Seicean A, Petrusel L, Seicean R. New targeted therapies in pancreatic cancer. World J Gastroenterol. 2015;21(20):6127–6145. doi:10.3748/wjg.v21.i20.6127
3. Akinleye A, Iragavarapu C, Furqan M, Cang S, Liu D. Novel agents for advanced pancreatic cancer. Oncotarget. 2015;6(37):39521–39537. doi:10.18632/oncotarget.3999
4. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–1617. doi:10.1056/NEJMra0901557
5. Chiorean EG, Coveler AL. Pancreatic cancer: optimizing treatment options, new, and emerging targeted therapies. Drug Des Devel Ther. 2015;9:3529–3545. doi:10.2147/DDDT.S60328
6. Cwik G, Wallner G, Skoczylas T. Cancer antigens 19-9 and 125 in the differential diagnosis of pancreatic mass lesions. Arch Surg. 2006;141(10):968–973. doi:10.1001/archsurg.141.10.968
7. Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J Gastrointest Oncol. 2012;3(2):105–119. doi:10.3978/j.issn.2078-6891.2012.005
8. Chen YZ, Liu D, Zhao YX. Diagnostic performance of serum macrophage inhibitory cytokine-1 in pancreatic cancer: a meta-analysis and meta-regression analysis. DNA Cell Biol. 2014;33(6):370–377. doi:10.1089/dna.2013.2237
9. Gold DV, Gaedcke J, Ghadimi BM, et al. PAM4 enzyme immunoassay alone and in combination with CA 19-9 for the detection of pancreatic adenocarcinoma. Cancer. 2013;119(3):522–528. doi:10.1002/cncr.27762
10. Zihao G, Jie Z, Yan L, et al. Analyzing S100A6 expression in endoscopic ultrasonography-guided fine-needle aspiration specimens: a promising diagnostic method of pancreatic cancer. J Clin Gastroenterol. 2013;47(1):69–75. doi:10.1097/MCG.0b013e3182601752
11. Koopmann J, Fedarko NS, Jain A, et al. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13(3):487–491.
12. Han W, Cao F, Chen MB, et al. Prognostic value of SPARC in patients with pancreatic cancer: a systematic review and meta-analysis. PLoS One. 2016;11:e0145803. doi:10.1371/journal.pone.0145803
13. Yang Y, Bai YS, Wang Q. CDGSH iron sulfur domain 2 activates proliferation and EMT of pancreatic cancer cells via Wnt/β-catenin pathway and has prognostic value in human pancreatic cancer. Oncol Res. 2017;25(4):605–615. doi:10.3727/096504016X14767450526417
14. Xia X, Wu W, Huang C, et al. SMAD4 and its role in pancreatic cancer. Tumour Biol. 2015;36(1):111–119. doi:10.1007/s13277-014-2883-z
15. Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt protooncogene product by phosphatidylinositol‐3,4‐bisphosphate. Science. 1997;275:
16. Li Chew C, Lunardi A, Gulluni F, et al. In vivo role of INPP4B in tumor and metastasis suppression through regulation of PI3K‐AKT signaling at endosomes. Cancer Discov. 2015;5(7):
17. Perez‐Lorenzo R, Gill KZ, Shen CH, et al. A tumor suppressor function for the lipid phosphatase INPP4B in melanocytic neoplasms. J Invest Dermatol. 2014;134(5):1359–1368. doi:10.1038/jid.2014.53
18. Hodgson MC, Shao L, Frolov A, et al. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Can Res. 2011;71(2):
19. Fedele CG, Ooms LM, Ho M, et al. Inositol polyphosphate 4‐phosphatase II regulates PI3K/Akt signaling and is lost in human basal‐like breast cancers. Proc Natl Acad Sci USA. 2010;107(51):
20. Guo ST, Chi MN, Yang RH, et al. INPP4B is an oncogenic regulator in human colon cancer. Oncogene. 2016;35(23):
21. Rijal S, Fleming S, Cummings N, et al. Inositol polyphosphate 4‐phosphatase II (INPP4B) is associated with chemoresistance and poor outcome in AML. Blood. 2015;125(18):
22. Chi MN, Guo ST, Wilmott JS, et al. INPP4B is upregulated and functions as an oncogenic driver through SGK3 in a subset of melanomas. Oncotarget. 2015;6(37):39891–39907. doi:10.18632/oncotarget.5359
23. Dzneladze I, Woolley JF, Rossell C, et al. SubID, a non-median dichotomization tool for heterogeneous populations, reveals the pan-cancer significance of INPP4B and its regulation by EVI1 in AML. PLoS One. 2018;13(2):e0191510. doi:10.1371/journal.pone.0191510
24. Zhang B, Wang W, Li C, Liu R. Inositol polyphosphate-4-phosphatase type II plays critical roles in the modulation of cadherin-mediated adhesion dynamics of pancreatic ductal adenocarcinomas. Cell Adh Migr. 2018;12(6):548–563. doi:10.1080/19336918.2018.1491496
25. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
26. Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:
27. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
28. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
29. Gewinner C, Wang ZC, Richardson A, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16(2):115–125. doi:10.1016/j.ccr.2009.06.006
30. Scheid MP, Huber M, Damen JE, et al. Phosphatidylinositol (3,4,5)P3 is essential but not sufficient for protein kinase B (PKB) activation; phosphatidylinositol (3,4)P2 is required for PKB phosphorylation at Ser-473: studies using cells from SH2-containing inositol-5-phosphatase knockout mice. J Biol Chem. 2002;277(11):9027–9035. doi:10.1074/jbc.M106755200
31. Chu LC, Goggins MG, Fishman EK. Diagnosis and detection of pancreatic cancer. Cancer J. 2017;23(6):333–342. doi:10.1097/PPO.0000000000000290
32. Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12(6):319–334. doi:10.1038/nrclinonc.2015.53
33. Du JD, Zheng X, Chen YL, et al. Elevated Transient Receptor Potential Melastatin 8 (TRPM8) expression is correlated with poor prognosis in panreatic cancer. Med Sci Monit. 2018;24:3720–3725. doi:10.12659/MSM.909968
34. Zhang JF, Hua R, Liu DJ, Liu W, Huo YM, Sun YW. Effect of CD74 on the prognosis of patients with resectable pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2014;13(1):81–86.
35. Yang JY, Jiang SH, Liu DJ, et al. Decreased LKB1 predicts poor prognosis in pancreatic ductal adenocarcinoma. Sci Rep. 2015;5:10575. doi:10.1038/srep10575
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.