Back to Journals » Journal of Hepatocellular Carcinoma » Volume 8
Inhibition of SIRT1 Limits Self-Renewal and Oncogenesis by Inducing Senescence of Liver Cancer Stem Cells
Authors Wang MJ , Chen JJ, Song SH, Su J, Zhao LH, Liu QG, Yang T, Chen Z, Liu C, Fu ZR, Hu YP, Chen F
Received 15 December 2020
Accepted for publication 2 June 2021
Published 29 June 2021 Volume 2021:8 Pages 685—699
DOI https://doi.org/10.2147/JHC.S296234
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmed Kaseb
Min-Jun Wang,1,* Jia-Jia Chen,1,* Shao-Hua Song,2,* Jing Su,1,* Ling-Hao Zhao,3 Qing-Gui Liu,1 Tao Yang,1 Zhiwen Chen,4 Chang Liu,4 Zhi-Ren Fu,2 Yi-Ping Hu,1 Fei Chen1
1Department of Cell Biology, Center for Stem Cell and Medicine, Second Military Medical University (Navy Medical University), Shanghai, People’s Republic of China; 2Department of General Surgery, Liver Transplantation Center, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China; 3National Center for Liver Cancer, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, People’s Republic of China; 4State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fei Chen Yi-Ping Hu
Department of Cell Biology, Center for Stem Cell and Medicine, Second Military Medical University, 800 Xiangyin Road, Shanghai, 200433, China
Tel +86-21-81870943
Email [email protected]; [email protected]
Purpose: Cancer stem cells (CSCs) have been considered involving in tumorigenesis, local recurrence, and therapeutic drug resistance of hepatocellular carcinoma (HCC). To investigate novel and effective methods for targeting hepatic CSCs is crucial for a permanent cure of liver cancer.
Methods: The expression level of SIRT1 was detected in CSCs of HCC tissues and cancer cell lines. Expression of CSC markers, the self-renewal and tumorigenic ability of liver CSCs were analyzed with SIRT1 inhibition. Cellular senescence-related markers were used to detect CSCs senescence after inhibition of SIRT1.
Results: SIRT1 was highly expressed in CSCs of HCC cell lines and human HCC tissues. In vitro study revealed that decreasing of SIRT1 level significantly downregulated the stemness-associated genes of liver CSCs and reduced the CSC stemness properties. Also, downregulated SIRT1 suppressed liver CSCs proliferation by decreasing their self-renewal abilities. Furthermore, CSCs with decreased SIRT1 expression showed limited tumorigenicity and formed smaller HCC tumor in vivo. And SIRT1 decreased CSCs became more susceptible to chemotherapeutic drugs. Mechanistically, SIRT1 decreased CSCs became senescence through the activation of p53-p21 and p16 pathway. The data further indicated that the tumor formed from SIRT1-knockdown CSCs exhibited higher senescence-associated β-galactosidase (SA-β-Gal) activity but lower proliferative capacity.
Conclusion: Taken together, these findings pointed that induction of senescence in liver CSCs is an effective tumor suppression method for HCC, and SIRT1 may be served as a promising target for HCC treatment.
Keywords: SIRT1, hepatocellular carcinoma, liver cancer stem cells, cellular senescence, stemness, self-renewal
Introduction
Hepatocellular carcinoma (HCC), as the most commonly primary liver tumor, is considered to be the sixth most prevalent malignancy and the second leading cause of cancer-related death worldwide.1,2 The incidence of HCC has been increasing in recent decades.3 In the past decade, due to recurrence and chemoresistance, 5-year survival rate of HCC patients remains at about 30%.4,5 In tumors, cancer stem cells (CSCs), also named tumor-initiating cells (TICs) are thought to have potential for self-renewal and could generate a variety of differentiated cells, and thus drive the tumorigenesis.6 Previous studies showed that CD13, CD133, EpCAM and other biomarkers could be used to identify liver CSCs.7–9 Accumulating evidence have showed that CSCs are less susceptible to chemotherapy and radiotherapy,10−12 which partly contribute to the recurrence of HCC in patients. Therefore, elucidation of the precise mechanisms underlying CSCs on HCC progression and development of novel therapeutic target for CSCs are essential for improving the current poor prognosis.
Recent study has been suggested that inducing cellular senescence might be a promising strategy for treating cancer.13 Cell cycle arrest is a typical characteristic of cellular senescence.14 Normal cells undergo senescence or death, and then are cleared by the body’s immune system. But tumor cells acquire mutations that cause them to escape from the normal cell cycle destiny.15 By intervening the cell cycle progression of tumor cells, senescence induction was considered as a potent protection for malignant transformation.16 Indeed, abundant evidence have indicated that cellular senescence is one of the tumor suppressor mechanisms.17–20 For example, inhibition of aspartate β-hydroxylase reduces human HCC cell proliferation by inducing cellular senescence,21 while knockdown of Arid1b promotes to HCC formation by blocking cell senescence.22 Hence, limiting capacity of self-renewal and differentiation by inducing CSCs senescence might be a more effective strategy to inhibit HCC development.
However, the underlying mechanisms between stem cells’ decline and CSC initiation are still ambiguous. NAD+-dependent deacetylase SIRT1, one of the most conserved longevity genes, is reported to participate in stress response, endocrine mechanism, aging, and cancer.23 As SIRT1 has been reported to act as a potential oncogene, it is usually overexpressed in various kinds of CSCs including breast cancer, colorectal cancer, leukemia and HCC. Furthermore, overexpression of SIRT1 is found to promote cancer development and is closely associated with poor prognosis in patients.24–28 Recent studies also demonstrate that SIRT1 overexpression is responsible for chemoresistance in cancers.29,30 Similarly, SIRT1 is also involved in aging and aging reversal of tissue-specific stem cells. For example, SIRT1-deletion caused a profound decline in hematopoietic stem cells (HSC) self-renewal and recapitulated the main properties of aged HSCs in young HSCs.31 And SIRT1 overexpression ameliorated aged mesenchymal stem cells (MSCs) senescent phenotype.32
Considering that CSCs in tumor play a vital for self-renewal and contribute to a new tumor generation, as well as the biological effect of SIRT1 on cancer and stem cell senescence, inhibition of SIRT1 expression might be a therapeutic intervention by inducing liver CSCs to senescence and alleviating the insensitivity to chemoresistance. In this study, we found that SIRT1 was highly expressed in the CSCs of human HCC tissues and HCC cell lines compared with the non-CSCs. Decreased SIRT1 level by short hairpin RNA (shRNA) intervention could significantly downregulate the stemness-associated genes and decrease the self-renewability of liver CSCs. Mechanistically, we identified that decreased SIRT1 level could induce CSCs senescence through activation of the p53-p21 and p16 pathway. Of note, liver CSCs with SIRT1 inhibition produced smaller HCC tumors in vivo and exhibited higher senescence-associated β-galactosidase (SA-β-Gal) activity. These findings indicated that the SIRT1'- mediated CSC senescence provided a molecular mechanism underlying the tumorigenic suppression of CSCs.
Materials and Methods
Cells and Tissue Samples
Human HCC cell lines (SMMC-7721, Huh7, Hep3B, LM3) were purchased from FuHeng Cell Center, Shanghai, China. HepG2 and 293T were purchased from ATCC, Virginia, US. SK-Hep1 and PLC/PRF5 were purchased from OBIO, Shanghai, China. Primary cultured human liver cancer cell line (CLC4 and CLC13) were gifts from Dr Hui Lijian’s Lab (CEMC, CAS, Shanghai, China) and maintained according to their published literature. HepG2, SMMC-7721, Huh7, Hep3B, LM3, PLC/PRF5 and 293T were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Grand Island, NY, US) supplemented with 10% fetal bovine serum (FBS; Gibco), penicillin (100 mg/mL) and streptomycin (100 U/mL) (Gibco). Human HCC cell line SK-Hep1 were cultured in Minimum Essential Medium (MEM; Gibco) supplemented with 10% FBS, 100 mg/mL penicillin and 100 U/mL streptomycin. CLC4 and CLC13 were maintained in RPMI1640 (Gibco) with 10% FBS, 1mM sodium pyruvate, 10 μg/mL insulin, 5.5 μg/mL transferrin, 40 ng/mL epidermal growth factor (EGF), and 6.7 ng/mL sodium selenite (Gibco). All cells were grown in an atmosphere containing 5% CO2 at 37°C. Human liver cancer specimens were obtained from the Eastern Hepatobiliary Surgery Hospital/Institute (Shanghai, China), with approval of the Institutional Review Committee of Shanghai Eastern Hepatobiliary Surgery Hospital and written informed consent of all patients.
Lentiviral Production and Infection
Lv-PNanog-GFP lentivirus was purchased from OBiO Technology (Shanghai). For Lentivirus-based shRNA knock-down systems, pLKO.1 is a replication-incompetent lentiviral vector chosen by the TRC for expression of shRNAs under the control of the human U6 promoter. The SIRT1-specific shRNA targeting sequences were purchased from Sigma-Aldrich. The oligonucleotide sequences of shRNA are: shSIRT1-1: 5ʹ-TACGATACAAGGCTGTTAGAGAG-3ʹ; shSIRT1-2: 5ʹ-CAGGTCAAGGGATGGTATTTA-3ʹ; shSIRT1-3: 5ʹ-CATGAAGTGCCTCAGATATTA-3ʹ; shSIRT1-4: 5ʹ-GCGGCTTGATGGTAATCAGTA-3ʹ; shScrambled: 5ʹ-CGTACGCGGAATACTTCGA-3ʹ. The oligos were cloned into the pLKO.1 TRC-cloning vector according to the Addgene’s pLKO.1 protocol. The lentivirus was produced by transfecting shRNA plasmid, lentiviral packaging plasmids psPAX2 and pMD2.G into HEK-293T cells with PEI transfection method. The medium containing the lentivirus was harvested by filtration after 48 and 72 hours of transfection. For lentiviral infection, cells were firstly plated into 6-well plates at 5×104 cells per well for 24–36h with a density of 50–70%, 100 μL lentivirus‐PNanog‐GFP or lentivirus‐shSIRT1 with 1×108TU/mL were respectively added in media and then replace with the cell media containing the viral mix. After 24h infection, virus-containing media was removed and replaced with normal media. Following infection, cells taking up virus were selected using 10 μg/mL puromycin, and lentiviral infected cells were analyzed by the next experiments.
RNA Isolation, RT-PCR and Quantitative PCR
Total RNA was extracted from cultured tumor cells, using the RNAiso Plus (Takara, Dalian, China) according to the manufacturer’s instructions. All reverse-transcriptase reactions were carried out with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Quantitative PCR was performed using primers and reagents on LightCycler 480 (Roche, Indianapolis, IN, USA). All samples were run in triplicate. Fold change was calculated by the 2-ΔΔCT method.33
Western Blot
Cell or tissue lysates were made in RIPA buffer supplemented with protease inhibitors cocktail and PMSF. Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Temecula, CA, US). Membranes were blocked in 5% milk/TBST 1h at the room temperature and then incubated in primary antibodies (Table S1) overnight at 4°C. After being washed by TBST three times, the membranes were incubated with HRP-conjugated secondary antibodies at 37°C for 30 minutes. Proteins were detected by PierceR ECL Western Blotting Substrate (Thermo Fisher, Waltham, MA, US).
Immunohistochemistry (IHC) and Immunocytochemistry (ICC)
For IHC staining, paraffin sections (2μm-thick) were de-paraffinized with xylene and ethanol, and incubated in 0.01 M citrate buffer (pH 6.0) at 121°C/100 kpa for 3 minutes to retrieve antigen. Samples were incubated with primary antibodies (Table S1) at 4 °C overnight, and then washed by PBS three times and incubated with fluorescent dye- or HRP-conjugated secondary antibody at 37 °C for 30 minutes. Nuclei were labeled with Hoechst 33,342 for fluorescent dye. DAB (Thermo Fisher) was used for HRP-conjugated antibody followed by counterstaining with hematoxylin (Sigma-Aldrich, St. Louis, MO, US) and covering in neutral balsam (Solarbio, Beijing, China).
For ICC staining, monolayer cells grown on coverslips were fixed with 4% PFA, and then permeabilized with 0.3% Triton X-100. Followed by block with 1% BSA at room temperature for 20–30 minutes. After that, cells were incubated with primary antibodies (Table S1) at 4 °C overnight, and then incubated with fluorescent dye-conjugated secondary antibody for 30 minutes at 37 °C. Nuclei were labeled with DAPI.
EdU Proliferation Assay
Cells were seeded in 24-well plates with cell slides. Cell proliferation was detected using the EdU Cell Proliferation Assay Kit (Invitrogen). According to the manufacturer’s protocol, the cells were incubated with EdU for 1–2 h before fixation, permeabilization and staining. The proportion of cells that incorporated EdU was determined by fluorescence microscopy.
Colony Formation Assays
For clone formation efficiency assay, 600 HepG2 cells or 900 SMMC-7721 cells were plated per well on six-well plates with culture medium for 2 weeks. Then, the clones were fixed by 4% formaldehyde and dyed with crystal violet and clone numbers (≥50 cells) were counted microscopically.
Sphere Formation Assays
For sphere formation efficiency assay, a total of 5000 cells were seeded into ultra-low attachment six-well plates (BD Biosciences, San Jose, CA, USA). Cells were cultured in DMEM/F12 supplemented with B27 (Gibco), N2 (Gibco), 20 ng/mL EGF and 20 ng/mL basic fibroblast growth factor (bFGF), 10 ng/mL hepatocyte growth factor (HGF) and 1% methyl cellulose (Sigma-Aldrich) was added to prevent cell aggregation. Cells were cultured in 37°C f incubator with 5% CO2 for 2 weeks, and the numbers of spheres (diameter >75μm) were counted.
SA-β-Gal Activity
Fix the cells planted on the six-well plates or the frozen tissue slices in advance using the fixative solution for 10 minutes according to the manufacturer’s instructions. After being washed with PBS, cells or tissue slices were stained with SA-β-Gal kit (Abcam, Cambridge, UK) at 37°C. Microscopic observation of cells stained blue, the percentage of which reflects the number of senescence-associated cells. For frozen tissue sections, the nucleus was stained with nuclear fast red.
Animal Studies
All animal experiments were performed in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. The study was approved by Animal Care and Welfare Committee of Second Military Medical University. For tumor formation assay, 102, 103, 104 cells mixed respectively with Matrigel at ratio of 1:1 and injected subcutaneously into the nude mice, the mice were sacrificed 1–2 months after inoculation. Tumors were harvested, counted and photographed.
Statistical Analysis
All data were presented as the mean ± SD. Statistical Analyses were conducted by GraphPad Prism 6.0. The Student’s t test was used when two groups were compared. P < 0.05 was considered statistically significant and is marked with “*”. P < 0.01 was considered highly statistically significant and is marked with “**”.
Results
SIRT1 is Highly Expressed in Liver CSCs of Human HCC Specimens and Cell Lines
Consistent with the previous studies,27 immunohistochemistry and Western blot assays revealed that SIRT1 expression was significantly elevated in tumor tissues compared with paired normal liver tissues in most patients (Figure 1A-C). Notably, we observed a strong distinct staining for SIRT1 with predominant nuclear location (Figure 1C) in HCC tissues. As known that liver CSCs closely contribute to HCC recurrence and chemoresistance, we analyzed the expression of SIRT1 in liver CSCs. Two surface markers of CSCs were co-staining with SIRT1 in HCC tissues. As shown in Figure 1D and 1E, SIRT1 was highly expressed in CD13 and EpCAM-positive cells. And we found that more than 60% CD13 or EpCAM-positive cells expressed SIRT1, while less than 15% CD13 or EpCAM negative cells expressed SIRT1 (Figure 1F). These results indicated that the expression of SIRT1 was positively correlated with CSCs of HCC.
Moreover, there was universally higher expression of SIRT1 in most human HCC cell lines (HepG2, CLC13, Hep3B, LM3, SMMC-7721, and SK-Hep1) (Figure 2A). Among these hepatoma cell lines, HepG2 and SMMC-7721 were chosen for subsequent experiments. To demonstrate whether SIRT1 was overexpressed in liver CSCs of HCC cell lines, we used the lentiviral fluorescent reporter system (Lv-PNanog-GFP) (Figure 2B) to isolate liver CSCs (NanogPos cells) and non-CSCs (NanogNeg cells) from HCC cell lines (HepG2 and SMMC-7721) (Figure 2C).34 Stemness-associated genes were analyzed by qPCR and the data showed that the levels of Nanog, OCT4, SOX2, CD13, and EpCAM were higher in sorted NanogPos cells than that in NanogNeg cells (Figure 2D). Western blotting demonstrated that the expression level of SIRT1 was obviously increased in liver CSCs than that in non-CSCs (Figure 2E and F). Meanwhile, immunofluorescence staining also demonstrated that SIRT1 was higher expressed in NanogPos liver CSCs than that in NanogNeg cells (Figure 2G). These data confirmed that SIRT1 was overexpressed in human HCC cell lines, especially in NanogPos liver CSCs.
Inhibition of SIRT1 Expression Decreases the Stemness Properties in Liver CSCs
To investigate the effect of SIRT1 on liver CSCs function, shRNA was used and SIRT1-knockdown hepatoma stem cells were constructed. We designed four shRNAs that target SIRT1, and found shSIRT1-2 was the most efficient one for inhibition of SIRT1 expression by Western blotting analysis (Figure 3A). Firstly, the determination of NANOG by flow cytometry indicated the decreased NanogPos liver CSCs after SIRT1 inhibition (Figure 3B). Then, the mRNA expression levels of pluripotent transcription factors and surface markers of CSCs (including NANOG, SOX2, OCT4, CD13, CD44 and EpCAM) were detected by qPCR, and we found that the stemness-associated transcription factors and CSC surface markers were significantly downregulated with SIRT1 knockdown (Figure 3C). Immunofluorescence staining indicated that the proportion of EpCAM-positive cells was also significantly reduced in SIRT1-silenced CSCs (Figure 3D). These findings demonstrated that decreasing SIRT1 level in liver CSCs strongly reduced the stem-like phenotype.
SIRT1 Knockdown Suppresses the Self-Renewal of Liver CSCs
To test the biological functions of SIRT1 on the self-renewal capacity of liver CSCs, we first performed the EdU incorporation assay to detect the cell proliferation. The result showed that SIRT1 knockdown significantly reduced the number of S-phase CSCs (Figure 4A and B). CSCs have the ability to form large colonies with a single cell, and this clonogenic activity is considered as an important indicator of undifferentiated state.35 In agreement with this notion, our data showed that accompanied with the lower proliferative capacity, the size and number of colonies were decreased in SIRT1-knockdown CSCs (Figure 4C and D). To further investigate the effect of SIRT1 on CSC self-renewal, sphere formation assay was performed. As shown in Figure 4E and 4F, in vitro spheroid formation assay showed that repression of SIRT1 level in NanogPos liver CSCs could significantly suppress the capacity of spheroid formation. Furthermore, sensitivity to sorafenib and cisplatin of SIRT1-inhibition in CSCs of HepG2 and SMMC-7721 was examined (Figure 4G and H). For shScrambled groups, the sorafenib IC50 values were (18.16 ± 0.40 μM) and (20.82 ± 0.53 μM), respectively; the cisplatin IC50 values were (51.81 ± 1.08 μg/mL) and (34.54 ± 0.85 μg/mL), respectively. For SIRT1-inhibition groups, the sorafenib IC50 values of HepG2 and SMMC-7721 CSCs were (11.91 ± 0.42 μM) and (15.42 ± 0.35 μM), respectively; the cisplatin IC50 values were (13.92 ± 0.36 μg/mL) and (9.54 ± 0.23 μg/mL), respectively. These data suggested that repression of SIRT1 in liver CSCs leads to decreasing resistance to chemotherapeutic drugs.
SIRT1 Knockdown Suppresses the Tumorigenesis of Liver CSCs
Next, we explored the contribution of SIRT1 to the tumorigenicity of liver CSCs by applying the subcutaneous tumor formation model of nude mice. In vivo limited dilution assay revealed that compared to implanted with the scramble CSCs, the number of animals that developed tumors was markedly reduced when implanted with the same numbers of SIRT1-silenced CSCs (Figure 5A). For example, 83.3% of the mice grew tumors when 1000 SMMC-7721 CSCs were implanted, but there was only 33.3% of mice developed tumors when implanted with SIRT1 silenced CSCs (Figure 5A). And the tumor size from the SIRT1 silencing group was visibly smaller than that from the control group (Figure 5B). Similar results were obtained in HepG2 stem cells with SIRT1 knockdown. In addition, we performed immunohistochemistry staining of the forming tumor tissues which was removed from mice. The results showed that the tumor tissues generated from liver shSIRT1 CSCs group exhibited lower SIRT1 levels (Figure 5C and D). Similarly, with SIRT1 knockdown, the EpCAM-positive CSCs were significantly reduced in the formed tumor after CSCs implantation (Figure 5E and F). Taken together, SIRT1 knockdown of CSCs inhibited their capacity of self-renewal and tumorigenic potential.
Liver CSCs Become Cellular Senescence via Activating p53-p21 and p16 Pathways After SIIT1 Inhibition
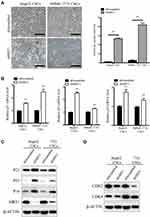
SIRT1 has been reported to be involved in aging and cellular senescence, but it is also considered as an effective mechanism of tumor suppression.36–39 We hypothesized that CSCs senescence induced by SIRT1 inhibition might be account for the decreased capacity of liver CSCs self-renewal and tumor formation. To further confirm this hypothesis, SA-β-Gal activity was used to investigate whether CSCs became senescence with SIRT1 knockdown. We found that SA-β-Gal activity was significantly increased in CSCs from two HCC cell lines after stably transfected with shSIRT1 hairpins (Figure 6A). We also observed that the senescent cells became enlarged and flat (Figure 6A). The senescence program can be initiated by multiple factors, such as cell cycle inhibitor proteins p53, p21, and p16Ink4a,40a,41 QPCR and Western blotting assay showed the expression of p21, p53 and p16 was significantly increased upon SIRT1 silencing in all two liver CSCs (Figure 6B and C). Meanwhile, we examined the levels of p53-p21 and p16 pathway-related proteins including CDK2 and CDK4. The data showed that the expression levels of CDK2 and CDK4 were decreased when inhibiting SIRT1 (Figure 6D). These findings implied that downregulated SIRT1 induced cellular senescence might be the main mechanism underlying the decreased self-renewal and suppressed tumorigenicity of liver CSCs. Moreover, the canonical p53-p21 and p16 signaling pathways was activated during cellular senescence.
Next, the tumor tissues formed with control and shSIRT1 CSCs implantation were analyzed with senescent and proliferative markers. The number of SA-β-Gal activated cells in the formed tumor was significantly increased in the SIRT1 silencing group when compared with the untreated group (Figure 7A). Moreover, a similar result was found by P21 staining (Figure 7B and C). However, a decreased expression of Ki67 was observed in tumor sections from shSIRT1 CSCs implantation (Figure 7D and E). These results revealed that SIRT1 knockdown induces the liver CSCs senescence and inhibits the cell growth in vivo.
Discussion
Frequent recurrence and therapeutic resistance are considered as a significant obstacle of HCC treatment, both of which was owing to the activation of CSCs. And liver CSCs have been reported to play an important role in maintaining hepatic tumorigenic properties.42 Thus, understanding of the molecular mechanisms on CSC initiation for tumor recurrence could be of great clinical benefit. SIRT1 has been reported to serve as an oncogene in HCC, which contributes to tumor cell growth and tumorigenesis.43 Moreover, SIRT1 is reported to contribute to maintain self-renewal and differentiated capacity of HSCs, especially under cellular stress progress.44 It has been reported that the SIRT1= mediated b-catenin overexpression promoted liver CSCs self-renewal and tumorigenesis.45 However, our study revealed that decreased the expression of SIRT1 could significantly induce liver CSCs became senescence, which was the key mechanism for the suppression of liver CSCs’ self-renewal and oncogenesis. We showed that expression of SIRT1 was co-localized with stem markers including EpCAM and CD13 in HCC tissues, which suggested a positive correlation between SIRT1 and liver CSCs biological function.
It has been reported that several surface markers including CD133, CD90, CD13, CD24, and EpCAM were used to isolate the CSCs from HCC.46–50 However, the exact biomarkers of CSCs and how they participated in the biological functions of CSCs in HCC is still unclear. The transcription factor Nanog is reported to contribute to maintain the stemness features of embryonic stem cells. Its expression is observed in germ cell induced tumors, embryonic carcinoma, and HCC.49 Of note, Nanog level is observed to significantly increased in CSC group, and Nanog displays a high correlation with tumor formation and stem cell behavior.49,51 Furthermore, recent studies have shown that Nanog-positive cancer cells not only express the stem cell markers, but also have the ability of self-renewal, colony formation, and differentiation for generating a new tumor.27,34 Therefore, lentiviral reporter system (Lv-PNanog-GFP) used as described can isolate the liver CSCs. In the study, we found that SIRT1 was higher expressed in NanogPos stem cells while that was in a lower level in NanogNeg cells. However, the stemness properties of NanogPos liver CSCs from HCC attenuated after decreasing the SIRT1 level. Inhibition of SIRT1 in CSCs also reduced expression of stemness-associated genes, such as OCT4, SOX9, CD13, CD24, and EpCAM. Further study indicated that SIRT1 knockdown significantly reduced the CSCs’ capacity to self-renew. In vivo tumor formation decreased when implanted with SIRT1 silencing liver CSCs, implying the potential decrease of HCC recurrence owing to inhibition of CSC self-renewal and tumorigenicity with SIRT1 knockdown.
One important reason for hepatocyte carcinoma development is that pre-malignant hepatocytes escape normal cell cycle arrest.37,52 Cellular senescence could be considered as a key mechanism to block proliferation of tumorigenic cells.37,52,53 Therefore, switching the quiescent or proliferative CSCs to senescent state will have potential clinical applications in improving the survival of HCC. Our results showed that the SIRT1 knockdown increased the CSCs SA-β-Gal activity and cell cycle arrest with feature of senescence. Activation of p53/p21 and p16 pathways was the molecular mechanism involved in cellular senescence.40,41 And we confirmed that inhibition of SIRT1 activated the expression of p53, p21, and p16. Furthermore, SIRT1 knockdown liver CSCs produced smaller HCC tumors in vivo but expressed higher SA-β-Gal activity. Therefore, SIRT1 intervention mediated cellular senescence provides a molecular mechanism underlying the tumorigenic suppressor of CSCs.
In summary, we showed that high expression of SIRT1 was found in liver CSCs of clinical human HCC specimens. Then, inhibition of SIRT1 expression deduced the stemness of liver CSCs, decreased several stemness-associated genes, as well as decreased the self-renewal capacity. SIRT1 silencing CSCs suppressed their tumorigenesis in vivo and limited the tumor growth. Mechanistically, inhibition of SIRT1 induced CSC senescence via the p53-p21 and p16 pathway activation. Our work highlights reduced SIRT1 expression promotes senescence of liver CSCs to inhibit their self-renewal and tumorigenesis, which is a potential strategy for decreasing HCC recurrence.
Abbreviations
CSCs, cancer stem cells; HCC, hepatocellular carcinoma; TICs, tumor-initiating cells; HSC, hematopoietic stem cells; MSCs, mesenchymal stem cells; shRNA, short hairpin RNA; SA-β-Gal, senescence-associated β-galactosidase.
Statement of Ethics
The use of human cell lines was approved by the Committee on Ethics of Medicine, Navy Medical University. Human specimens were obtained and employed with approval of the ethical committee of Shanghai Eastern Hepatobiliary Surgery Hospital and was conducted in accordance with the Declaration of Helsinki.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0107500), National Natural Science Foundation of China (31601101, 31701186, 32070732, 31970753), Medical Science Youth Development Plan of PLA (17QNP011), Open Research Fund of State Key Laboratory of Genetic Engineering, Fudan University (No. SKLGE1901).
Disclosure
The authors declare no conflicts of interest.
References
1. Gravitz L. Liver cancer. Nature. 2014;516(7529):S1. doi:10.1038/516S1a
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
3. Adnane L, Trail PA, Taylor I, Wilhelm SM. Sorafenib (BAY 43-9006, Nexavar), a dual-action inhibitor that targets RAF/MEK/ERK pathway in tumor cells and tyrosine kinases VEGFR/PDGFR in tumor vasculature. Methods Enzymol. 2006;407:597–612.
4. Shah C, Mramba LK, Bishnoi R, Bejjanki H, Chhatrala HS, Chandana SR. Survival differences among patients with hepatocellular carcinoma based on the stage of disease and therapy received: pre and post sorafenib era. J Gastrointest Oncol. 2017;8:789–798.
5. Wörns MA, Galle PR. HCC therapies–lessons learned. Nat Rev Gastroenterol Hepatol. 2014;11:447–452.
6. Clara JA, Monge C, Yang YZ, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells- a clinical update. Nat Rev Clin Oncol. 2020;17:204–232.
7. Yamashita T, Ji J, Budhu A, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024.
8. Haraguchi N, Ishii H, Mimori K, et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120:3326–3339.
9. Ma S, Tang KH, Chan YP, et al. miR-130b promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707.
10. Chen J, Jin R, Zhao J, et al. Potential molecular, cellular and microenvironmental mechanism of sorafenib resistance in hepatocellular carcinoma. Cancer Lett. 2015;367:1–11.
11. Li Y, Laterra J. Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer Res. 2012;72:576–580.
12. Zhou G, Latchoumanin O, Bagdesar M, et al. Aptamer-based therapeutic approaches to target cancer stem cells. Theranostics. 2017;7:3948–3961.
13. Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705.
14. van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509:439–446.
15. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70.
16. He S, Sharpless NE. Senescence in Health and Disease. Cell. 2017;169:1000–1011.
17. Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496.
18. He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169:1000–1011.
19. Braig M, Schmitt CA. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 2016;66:2881–2884.
20. Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21:1424–1435.
21. Iwagami Y, Huang CK, Olsen MJ, et al. Aspartate β-hydroxylase modulates cellular senescence through glycogen synthase kinase 3β in hepatocellular carcinoma. Hepatology. 2016;63:1213–1226.
22. Tordella L, Khan S, Hohmeyer A, et al. SWI/SNF regulates a transcriptional program that induces senescence to prevent liver cancer. Genes Dev. 2016;30:2187–2198.
23. Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295.
24. Ma W, Xiao GG, Mao J, et al. Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget. 2015;6:10432–10444.
25. Chen X, Sun K, Jiao S, et al. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci Rep. 2014;4:7481.
26. Li L, Osdal T, Ho Y, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014;15:431–446.
27. Liu L, Liu C, Zhang Q, et al. SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells. Hepatology. 2016;64:814–827.
28. Zhang S, Huang S, Deng C, et al. Co-ordinated overexpression of SIRT1 and STAT3 is associated with poor survival outcome in gastric cancer patients. Oncotarget. 2017;8:18848–18860.
29. Mvunta DH, Miyamoto T, Asaka R, et al. SIRT1 regulates the chemoresistance and invasiveness of ovarian carcinoma cells. Transl Oncol. 2017;10:621–631.
30. Chen H, Zhang W, Cheng X, et al. β2-AR activation induces chemoresistance by modulating p53 acetylation through upregulating Sirt1 in cervical cancer cells. Cancer Sci. 2017;108:1310–1317.
31. Rimmelé P, Bigarella CL, Liang R, et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Rep. 2014;3:44–59.
32. Liu X, Chen H, Zhu W, et al. Transplantation of SIRT1-engineered aged mesenchymal stem cells improves cardiac function in a rat myocardial infarction model. J Heart Lung Transplant. 2014;33:1083–1092.
33. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108.
34. Shan J, Shen J, Liu L, et al. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology. 2012;56:1004–1014.
35. Rajendran V, Jain MV. In vitro tumorigenic assay: colony forming assay for cancer stem cells. Methods Mol Biol. 2018;1692:89–95.
36. Di Mitri D, Alimonti A. Non-cell-autonomous regulation of cellular senescence in cancer. Trends Cell Biol. 2016;26:215–226.
37. Kang TW, Yevsa T, Woller N, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551.
38. Xu C, Wang L, Fozouni P, et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat Cell Biol. 2020;22:1170–1179.
39. Ong ALC, Ramasamy TS. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res Rev. 2018;43:64–80.
40. Ou HL, Schumacher B. DNA damage responses and p53 in the aging process. Blood. 2018;131:488–495.
41. Uyar B, Palmer D, Kowald A, et al. Single-cell analyses of aging, inflammation and senescence. Ageing Res Rev. 2020;64:101156.
42. Chiba T, Iwama A, Yokosuka O. Cancer stem cells in hepatocellular carcinoma: therapeutic implications based on stem cell biology. Hepatol Res. 2016;46:50–57.
43. Chen J, Zhang B, Wong N, et al. Sirtuin 1 is upregulated in a subset of hepatocellular carcinomas where it is essential for telomere maintenance and tumor cell growth. Cancer Res. 2011;71:4138–4149.
44. Singh SK, Williams CA, Klarmann K, Burkett SS, Keller JR, Oberdoerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J Exp Med. 2013;210:987–1001.
45. Chen X, Huan H, Liu C, et al. Deacetylation of beta-catenin by SIRT1 regulates self-renewal and oncogenesis of liver cancer stem cells. Cancer Lett. 2019;463:1–10.
46. Zhou G, Da Won Bae S, Nguyen R, et al. An aptamer-based drug delivery agent (CD133-apt-Dox) selectively and effectively kills liver cancer stem-like cells. Cancer Lett. 2021;501:124–132.
47. Yang ZF, Ho DW, Ng MN, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166.
48. Hu B, Xu Y, Li YC, et al. CD13 promotes hepatocellular carcinogenesis and sorafenib resistance by activating HDAC5-LSD1-NF-κB oncogenic signaling. Clin Transl Med. 2020;10:e233.
49. Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24+ liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63.
50. Nio K, Yamashita T, Okada H, et al. Defeating EpCAM(+) liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J Hepatol. 2015;63:1164–1172.
51. Chen CL, Uthaya Kumar DB, Punj V, et al. NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 2016;23:206–219.
52. Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522.
53. Childs BG, Baker DJ, Kirkland JL, Campisi J, van Deursen JM. Senescence and apoptosis: dueling or complementary cell fates? EMBO Rep. 2014;15:1139–1153.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.