Back to Journals » Drug Design, Development and Therapy » Volume 17
Informatics on Drug Repurposing for Breast Cancer
Authors Zhou H, Liu H, Yu Y, Yuan X , Xiao L
Received 17 April 2023
Accepted for publication 17 June 2023
Published 28 June 2023 Volume 2023:17 Pages 1933—1943
DOI https://doi.org/10.2147/DDDT.S417563
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Hui Zhou,1,2,* Hongdou Liu,3,* Yan Yu,3 Xiao Yuan,3,4 Ling Xiao5
1Department of Lymphoma and Hematology, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, People’s Republic of China; 2Department of Lymphoma and Hematology, Hunan Cancer Hospital, Changsha, Hunan, People’s Republic of China; 3Department of Laboratory Diagnosis, Changsha Kingmed Center for Clinical Laboratory, Changsha, Hunan, People’s Republic of China; 4Department of Laboratory Diagnosis, Guangzhou Kingmed Center for Clinical Laboratory, Guangzhou, Guangdong, People’s Republic of China; 5Department of Histology and Embryology of Xiangya School of Medicine, Central South University, Changsha, Hunan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ling Xiao, Department of Histology and Embryology of Xiangya School of Medicine, Central South University, Changsha, Hunan, People’s Republic of China, Tel +86-13974871376, Email [email protected] Xiao Yuan, Changsha Kingmed Center for Clinical Laboratory, Changsha, Hunan, People’s Republic of China, Tel +86-18662520739, Email [email protected]
Abstract: Moving a new drug from bench to bedside is a long and arduous process. The tactic of drug repurposing, which solves “new” diseases with “old” existing drugs, is more efficient and economical than conventional ab-initio way for drug development. Information technology has dramatically changed the paradigm of biomedical research in the new century, and drug repurposing studies have been significantly accelerated by implementing informatics techniques related to genomics, systems biology and biophysics during the past few years. A series of remarkable achievements in this field comes with the practical applications of in silico approaches including transcriptomic signature matching, gene-connection-based scanning, and simulated structure docking in repositioning drug therapies against breast cancer. In this review, we systematically curated these impressive accomplishments with summarization of the main findings on potentially repurposable drugs, and provide our insights into the current issues as well as future directions of the field. With the prospective improvement in reliability, the computer-assisted repurposing strategy will play a more critical role in drug research and development.
Keywords: drug repurposing, breast cancer, CMap, network, molecular docking
Introduction
One of the ultimate missions of biomedical research is to connect human diseases with causal genes and effective drugs. The successful deciphering of the human genome sequence at the beginning of this century accelerated the elucidation of the associations between genes and diseases. However, drug development remains daunting due to the lengthy bench-to-bedside process and the extremely high cost and low success rate. Drug repurposing refers to developing approved and investigational chemicals or biologics for novel utilization beyond their initial medical indications. With compounds having existing research basis and risk elimination, this “drug-recycling” approach could significantly reduce the time and cost of introducing novel drugs. Previous advancements, including successful drug repurposing examples and late-stage clinical candidates arising from deliberate repurposing studies, proved the feasibility and reliability of this tactic.1
Breast cancer (BC) is a heterogeneous disease that occurs in the epithelial tissue of the breast and is the leading cause of cancer death among female cohorts, with approximately 627,000 died cases worldwide in 2018.2 Although management for certain BC subtypes has been greatly improved over the past decades, and up to 30 BC drugs received the Food and Drug Administration (FDA) approval between 2010 and 2020, there is a continued need for the development of therapies for patients with treatment-resistant or refractory disease.3 Besides, some subtypes, such as triple-negative breast cancer (TNBC), still lack targeted therapies.4 Therefore, as a shortcut to drug discovery, drug repurposing has already become an essential supplementary means for BC drug developers,5 and more than a dozen medicines have been approved for BC treatment through this method.6
Repositioning an old drug for a specific disease was estimated to save 3–5 years compared to de novo drug development due to the well-defined pharmacodynamics and pharmacokinetics, as well as known side effects and metabolic profiles.5 In the past, some redirected drugs were discovered by accidental findings in the retrospective clinical analyses.1 For example, Minoxidil, once for hypertension on the market, was identified with an adverse effect of hair growth and is now a popular treatment for pattern hair loss and baldness. Several other reprofiled drugs were spotted via experimental approaches, including pharmacological assay and in vitro screening of compound libraries.1 Fingolimod was the first oral disease-modifying therapy approved for multiple sclerosis. The initial indication of this drug was transplant rejection, and the new use was revealed by pharmacological and structural analysis.
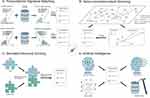
Either occasional observations or labor-intensive experimental screenings are often not hypothesis-driven.7 In contrast, pinpointing a drug with (1) a specific transcriptomic perturbative effect (Figure 1A), (2) a certain target connected to the undruggable gene/gene to be drugged in a pathway/network (Figure 1B), or (3) designated docking ability and binding stability (Figure 1C), through in silico way is more directional. In addition to this strong purposiveness, the high-throughput-and-low-cost feature makes the computational approaches promising in substantively facilitating drug discovery and availability. In this review, we focused on and summarized the advances in the innovative methodology driven by advanced informatics for BC drug repurposing with the curation of main findings on potentially repurposable drugs, and discussed the associated limitations and future directions in the field. We sincerely hope our review could bring clues and inspiration to the practitioners devoted to BC drug discovery.
Transcriptomic Signature Matching
Theoretically, when two drugs lead to a similar gene expression profile alteration, they might have a therapeutic effect on the same disease. Here, a study guided by this forward transcriptomic signature matching theory (Figure 1A) was described first.
The transcription factor estrogen receptor α (ERα), a driver of cancer progression was expressed in about 70% of BC cases.8 ERα-positive BCs were treated with endocrine therapy (ET) agents that interfere with ERα signal. Busonero et al constructed a particular “estrogenic signature” downregulated by conventional ERα antagonists in the human breast cancer cell MCF-7.9 With the 57-gene signature, they screened more than 60,000 compounds in the Connectivity Map (CMap)10 to find drugs with a similar downward signature. A list of candidate drugs was generated by comparing the concordance of gene signatures. Three drugs including mitoxantrone, thioridazine and menadione were selected for the subsequent experimental validation and eventually, thioridazine was identified as the preferential one with the ability to induce ERα degradation and inhibit the proliferation of ERα-positive BC cells.
As with reverse transcriptomic signature matching, a drug inducing transcriptomic changes opposite to those in a disease is considered to hold the potential to therapeutically reverse the illness (Figure 1A). Reverse matching utilizes prioritized genes from the result of differential gene expression analysis of disease transcriptomic data. Generally, top and bottom-ranked genes are selected as the signature of the disease. This transcriptomic signal is then compared with that generated from the expression profiles of drug interferences. Finally, the degree of negative correlation determines the drug usability and priority.
Vasquez-Bochm et al compared a reported signature containing 25 upregulated and 14 downregulated genes of breast cancer stem cell (BCSC), to the cell-line transcriptomic changes induced by over 1300 compounds in CMap.11 They identified five candidate drugs with reversed signals. Subsequent in vitro experiments confirmed that lovastatin was an effective BCSC-targeting drug by inhibiting SOX2 promoter transactivation and reducing mammosphere formation efficiency.
CMap10 for signature matching query is a famous cloud-based computing infrastructure with a database containing over one million gene expression profiles from thousands of compounds and reagents tested in multiple cell types. Thanh et al proposed DeCoST (Drug Repurposing from Control System Theory) based on the system control paradigm to tackle the drug repurposing tasks, and compared its performance with CMap.12 Before matchmaking, genes overexpressed and under-expressed in disease were marked by 1 and −1 respectively, while genes that were known to be activated and inhibited by a drug were quantified as +1 and −1 separately. Via reverse matching, DeCoST reprofiled eight drugs for ERα-positive BC and two for ERα-negative. The authors held that their work could provide complimentary capability to CMap for that DeCoST utilized tissue-level expression profiles while CMap used cell-line-level.
Appropriate query transcriptomic signal selection is crucial for the signature matching strategy. However, determining the optimal query signal is tricky in a real scenario, and the selection criteria vary drastically among studies.13 In order to solve this issue, Chan et al developed Dr Insight (Drug Repurposing: Integration and Systematic Investigation of Genomic High-Throughput data), which employed order statistics to directly measure the reverse association between disease and drug-perturbed expression data genome-wide.14 This automated tool eliminated the need for subjective selection of fixed-sized query signatures and identified five new drug candidates for BC using The Cancer Genome Atlas (TCGA) dataset.
BC is a highly heterogeneous disease, and genomic and transcriptomic characterization differ significantly among patients. This inter-tumor heterogeneity subdivides BC at the molecular level, and each subtype has unique treatment schedules. Mejía-Pedroza et al developed a pathway-based drug repositioning method for BC subtypes.15 Based on two public large-scale gene expression datasets, the authors found the most deregulated pathways for each subtype via a robust probabilistic manner. Each deregulated pathway was associated with its known pharmacological targets according to information from the Drug–Gene Interaction Database (DGIdb).16 These associations were finally classified to generate a contextual prioritization of pathways and drugs in BC subtypes. With this method, 79 drugs over eight significantly deregulated pathways displayed potentialities for transcriptomic recovery, and individualized drug regimens were provided for two patients with basal subtype.
Single-cell transcriptome sequencing technology has enabled the detailed characterization of intratumoral heterogeneity in BC.17,18 He et al invented a drug repurposing recommendation tool inspired by single-cell RNA data.19 This tool identified differentially expressed genes (DEG) between tumor and normal cells within each cell cluster (type), and then queried against its drug reference library to seek and rank compounds which significantly reversed the expression pattern in a single cluster or multiple clusters. The tool’s performance was validated with public TNBC data and several top-ranked drugs were found to have been approved by FDA or used in clinical trials.
Gene-connection-based Scanning
The development and progression of BC is a systematic process involving multiple interactions among different genes. Theoretically, a genotype-phenotype association could make the gene a prospective drug target for that altering the effect of the gene would contribute to achieve a desired clinical outcome.20 However, if the gene product, ie, a protein or an RNA molecule could not be perturbed via compounds, directly or indirectly connected genes upstream or downstream of the same pathway, or among the interactive network, could be chosen as alternative drug targets (Figure 1B). Through this compromise, existing drugs aimed at those connected genes could be explored for repurposing.
Protein–protein interaction (PPI) networks characterize physical interactions between proteins. Ma et al systematically investigated the cluster of PPI network of BC cell line MCF-7 by using four cluster detection algorithms and compared the performance of these algorithms for drug target prediction.21 The authors revealed that the Walktrap (CW) cluster detection algorithm performed best in extracting functional clusters from the network. By integrating the extracted clusters produced by CW with drug-induced differential gene expression data, potential drugs redirected to BC were provided. In this research, the authors pointed out that the connections in the PPI network should be modified by the transcriptomic landscape of specific cell lines, as the dynamic gene expression profiles among cell types caused differential functional interaction patterns between the molecular components. In another research, Turanli et al noted that different subtypes of BC had different PPI networks.22 They presented an integrated omics approach with transcriptome and interactome data to identify active PPI networks in TNBC patients. EED, DHX9, and AURKA were found to be aberrantly activated in TNBC tumors compared to both normal tissues and other BC subtypes, and thus proposed as potential drivers of proliferation as well as candidate drug targets. The authors queried each gene signature separately against the L1000CDS2 database23 and identified ten drugs with potentiality to reverse the aberrant activity in basal-like tumors.
Azam et al built a drug-disease network by considering all interactions between drug targets and disease-related genes in the context of all KEGG human-signaling pathways.24 A repurposing score was computed for each drug-disease pair by integrating expression data into the network. Finally, a ranked list of drugs with potential therapeutic effects for the given disease was generated based on the repurposing score. This method proposed six candidate drugs with preliminary evidence from preclinical or clinical studies against BC. Also with KEGG data, Firoozbakht et al developed a network-based integration approach to find drugs for each BC subtype.25 Copy number variation and aberration data were employed in their work for disease subtyping.
In optimization theory, the maximum flow problem is defined as finding a feasible flow through a flow network that obtains the maximum possible flow rate. In drug repurposing, the idea is that proteins with the maximum flow to disease-related risk proteins in a network could be regarded as an alternative drug target for the disease. Based on this theory, Islam et al constructed a maximum flow-based PPI network through which new drug targets were identified from the targets of the FDA drugs and their associated drugs for chronic disease treatment.26 Existing drugs were repurposed based on the maximum flow values of each newly identified target protein. The top four repurposed drugs for BC had been reported by other independent studies, demonstrating the feasibility of this framework.
A hub in a network refers to a core point which connects multiple elements and is therefore regarded as a vital network component. Neoadjuvant chemotherapy (NAC) was the frontline treatment for patients with locally advanced BC before tumor excision. However, drug resistance remained a major issue in NAC. Hence, identifying the key genes involved in driving NAC resistance and targeting them with approved drugs was a cost-effective solution. Detroja et al identified 1446 DEGs from public RNA-seq datasets of NAC-resistant BC patients.27 Subsequently, key hub genes were obtained via gene co-expression network analysis and Multiple Correlation Clustering. With four publicly available databases, they finally identified 19 prospective drugs, including eight FDA-approved ones for redirecting to the hub genes.
According to the drug-gene-disease relationship, a protein–protein network could evolve into a direct drug–drug network. Di et al28 developed an R-based package PriorCD for cancer drug repurposing by first considering the drug functional similarities at the pathway level. The research team constructed a functional similarity network between drugs by enriching mRNA and microRNA expression data into pathway activity profiles and correlating them with drug activity profiles derived from the drug–disease relationship. Drug prioritizing scores were calculated based on a global network propagation algorithm. The authors evaluated and validated the performance of this in silico approach by using several kinds of public resource. At last, 14 prioritized candidate drugs for BC were provided and four were FDA-approved.
Gene network could be coupled with other networks consisting of heterogeneous biomedical entities. Al-Taie et al dissected TNBC patients into five subgroups based on genotypic data and clinical information including age, race and neoplasm subdivision, and discovered appropriate therapeutic candidates separately through drug repurposing.29 In particular, this research team built a drug repositioning knowledge network which employed a gene-centric schema including relations between genes, pathways, GO domains, disease and drugs. With the curated knowledge base, the researchers repurposed drugs for each subgroup based on the gene expression patterns and the relationship between genes and other biomedical entities. They concluded that different targeted mechanisms should be suggested for different BC subgroups.
Simulated Structure Docking
Computationally predicting the complementarity of the binding area between a drug and a therapeutic target (Figure 1C) is another strategy for drug repositioning. This kind of method simulates the dynamic behavior of compound-target binding and predicts the stability of the binding compound by calculating the binding free energy. The recent progress using this structure-based virtual screening for BC drugs is briefly described here.
EGFR and HER2 were frequently overexpressed in BC and were targets of many approved anti-BC drugs.30 Balbuena-Rebolledo et al obtained dozens of FDA-approved drugs structurally similar to lapatinib and gefitinib, two well-known inhibitors of EGFR and HER2.31 These drugs were then investigated through a series of in silico prediction programs including virtual molecular docking, molecular dynamics (MD) simulation and the molecular mechanics generalized Born surface area (MM-GBSA) binding free energy prediction. Specifically, the best binding structures in the docking studies were chosen using the criteria of the lowest energetic ligand conformations at the receptor’s binding site. Then, the prevalence of the interactions was evaluated during MD simulation and unstable protein-ligand complexes were discarded. Binding free energy was calculated to measure the binding affinity, and compounds with higher affinities for EGFR/HER2 than those of known inhibitors were identified. In the in vitro assay, five drugs showed growth inhibitory activity on BC cell lines at micromolar concentrations. With the same series of in silico methods, Kandasamy et al predicted four compounds as potential repurposed drugs for target blocking in BC.32 Their in vitro studies demonstrated the anti-proliferative property of a known anti-psychotic drug Pimozide.
The Notch signaling pathway was a pivotal regulatory component in BC etiology and progression, and most associated genes were overexpressed in BC.33 One method of effectively blocking NOTCH activity was preventing its cleavage at the cell surface with γ-secretase inhibitors. Pathak et al used molecular docking analysis to virtually screen 1615 FDA-approved drugs and found that Venetoclax was superior in simulated docking capability to one of the standard γ-secretase inhibitors: RO4929097.34 The Venetoclax-γ-secretase complex was computationally confirmed in stability and thus was inferred with a probable inhibitory effect on NOTCH activity. Rui et al found that many γ-secretase inhibitors had various side effects, and impeding the Recombination Signal Binding Protein for Immunoglobulin Kappa J Region (RBPJ), an essential transcription factor in Notch signaling, could be a more specific way.35 The authors collected 10,527 pharmacologically active compounds by integrating four public database and used the docking module of Molecular Operating Environment, the GROMACS software package and the molecular mechanism/Poisson–Boltzmann surface area (MM-PBSA) binding free energy prediction for virtual screening of RBPJ inhibitors. Fidaxomicin, schaftoside and acarbose were identified as the most robust RBPJ binders for the strict occupation of the binding site and the formation of hydrogen bonds with multiple key RBPJ amino acid residues within the binding area. After cellular assay and in vivo anticancer investigation, FDA-approved fidaxomicin was identified as a potential RBPJ inhibitor against BC.
Drug repurposing strategies could also be employed in finding new indications for natural active compounds in clinical trials or still under investigation. Maruca et al explored the probable anticancer activities of native compounds from mushroom species.36 In their study, an in-house chemical database was utilized for the virtual screening against the isoform 7 of the Histone deacetylase (HDAC), a gene that regulated cellular proliferation, differentiation and development in BC. Via in silico docking simulation, MD simulation and cell viability assay, the authors proposed ibotenic acid as a lead compound for developing novel HDAC7 inhibitors.
Artificial Intelligence
Artificial intelligence (AI) has enabled leaps in discovering novel drug targets and drug-disease associations in multiple cancer types.37 Recently, AI technologies including machine learning, deep learning, neural network and data/text mining (Figure 1D) have been implemented in the investigations of BC drug repositioning.
Song et al developed a feature-based method to predict unknown drug–target interaction (DTI) called PsePDC-DTIs.38 The authors investigated seven common classification algorithms of machine learning and observed that random forest outperformed others in the quality of a prediction model. In exploring new targets for BC treatment using identified risk genes, the PsePDC-DTIs model provided ten potential drugs, six of which were found with direct or inferred evidence. Some traditional machine learning approaches showed limits in efficiently analyzing high-dimensional datasets extracted from drugs and targets. Given this, You et al proposed LASSO (least absolute shrinkage and selection operator)-based deep learning method to predict DTIs.39 The method successfully identified five BC drugs with literature evidence. As one form of deep learning, the neural network was also applied in drug reprofiling against BC. Chen et al proposed a graph neural network model called GraphRepur.40 The research team constructed a graph containing drug–drug links information and drug gene expression signatures. They established a drug repurposing prediction model by extracting the drug signatures and topological structure information in the graph. Among the predicted BC drugs by the model, ten had supported literature. Data/text mining is a process of finding and extracting patterns from massive datasets using computer programming, and it greatly facilitates knowledge discovery in many fields. Wang et al developed a recommender system named ANTENNA for the prediction of novel drug-gene-disease associations by mining large-scale chemogenomics and disease association data.41 With this system, the authors predicted Diazoxide for TNBC targeted therapy. Ji et al conducted text mining in about 3.8 million cancer-related PubMed abstracts and identified 18 diseases similar to inflammatory breast cancer (IBC). As a result, 24 drugs were proposed for redirection.42
Discussion
The journey of obtaining a safe and effective medicine to market is long and arduous, and only a few dozen drugs are licensed officially each year. Behind are tens of thousands of failed candidate drugs. In the researches collected in our review (Table 1), a limited part of candidate drugs redirected against BC succeeded in in-vitro validations (Table 1 Column 4 underlined), not to mention further in vivo experiments and rigorous preclinical/clinical trials with high rejection rates.43 One of the possible reasons is the uncertain predictive reliability by which general in silico strategies are always challenged. To be specific, first, the current reverse Transcriptomic Signature Matching manner (Figure 1A) commonly exploits disease signals generated from bulk RNA-seq data which reflects the expression profile of a cell mixture. However, not all these cells play equal roles in the disease or have a relationship to the ailment. Hence, the biased disease signal might lead to inaccurate drug matching. Second, the Gene-connection-based Scanning method (Figure 1B) is highly dependent on the existing knowledge base storing the information about the physical interactions between proteins (genes). Nevertheless, PPI databases contain a few false-positive interactions.44 This defect could bring uncertainty to the final predictive results. Third, for the Simulated Structure Docking approach (Figure 1C), neither of the current binding free energy calculation methods is reliable enough in drug-target binding affinity prediction.45
 |
Table 1 Brief Introduction and Candidate Drugs of Each Research Collected in This Review |
Here, for each of the three main tactics mentioned in our review, we suggest a possible solution or direction for the future improvement of predictive performance:
- Employing single-cell expression profile alteration data to pinpoint efficient drugs for particular cell cluster(s) allows for avoiding non-target cell clones.46
- Integration of a PPI network in a multi-omics context is conducive to highlighting key proteins or central clusters in drug action and response mechanisms, and removing the false interactions.47
- Cross-verification using multiple binding free energy calculation methods as all such methods currently have limitations and advantages that partially overlap and complement one another.45
This article summarized the recent progress of computational drug repurposing approaches in BC. Even though at the early stage of development, informatics-driven repurposing approaches are rational to serve as a preceding routine for preliminary drug screening prior to in vitro/in vivo assays. With the joint efforts of the whole community, this kind of repurposing strategy is promising in revolutionizing the present paradigm of drug research and development toward precision oncology.
Data Sharing Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Acknowledgment
This study was funded by the Scientific Research Project of Hunan Provincial Health Commission [202203045455]; the “Scientific Research Climbing Plan” of Hunan Cancer Hospital [ZX2020003]; and the Science and Technology Planning Project of Guangzhou [202011020002].
Disclosure
H.L., Y.Y. and X.Y. are all employees of Changsha Kingmed Center for Clinical Laboratory. X.Y. is also an employee of Guangzhou Kingmed Center for Clinical Laboratory. All other authors declare no financial or non-financial competing interests in this work.
References
1. Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi:10.1038/nrd.2018.168
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
3. Arora S, Narayan P, Osgood CL, et al. U.S. FDA drug approvals for breast cancer: a decade in review. Clin Cancer Res. 2022;28:1072–1086. doi:10.1158/1078-0432.CCR-21-2600
4. Yam C, Mani SA, Moulder SL. Targeting the molecular subtypes of triple negative breast cancer: understanding the diversity to progress the field. Oncologist. 2017;22:1086–1093. doi:10.1634/theoncologist.2017-0095
5. Aggarwal S, Verma SS, Aggarwal S, Gupta SC. Drug repurposing for breast cancer therapy: old weapon for new battle. Semin Cancer Biol. 2021;68:8–20. doi:10.1016/j.semcancer.2019.09.012
6. Malik JA, Ahmed S, Jan B, et al. Drugs repurposed: an advanced step towards the treatment of breast cancer and associated challenges. Biomed Pharmacother. 2022;145:112375. doi:10.1016/j.biopha.2021.112375
7. Mottini C, Napolitano F, Li Z, Gao X, Cardone L. Computer-aided drug repurposing for cancer therapy: approaches and opportunities to challenge anticancer targets. Semin Cancer Biol. 2021;68:59–74. doi:10.1016/j.semcancer.2019.09.023
8. Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi:10.1038/nrclinonc.2015.117
9. Busonero C, Leone S, Bianchi F, Acconcia F. In silico screening for ERalpha down modulators identifies thioridazine as an anti-proliferative agent in primary, 4OH-tamoxifen-resistant and Y537S ERalpha-expressing breast cancer cells. Cell Oncol. 2018;41:677–686.
10. Subramanian A, Narayan R, Corsello SM, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–52 e17. doi:10.1016/j.cell.2017.10.049
11. Vasquez-Bochm LX, Velazquez-Paniagua M, Castro-Vazquez SS, et al. Transcriptome-based identification of lovastatin as a breast cancer stem cell-targeting drug. Pharmacol Rep. 2019;71:535–544. doi:10.1016/j.pharep.2019.02.011
12. Nguyen TM, Muhammad SA, Ibrahim S, et al. DeCoST: a new approach in drug repurposing from control system theory. Front Pharmacol. 2018;9:583. doi:10.3389/fphar.2018.00583
13. Musa A, Ghoraie LS, Zhang SD, et al. A review of connectivity map and computational approaches in pharmacogenomics. Brief Bioinform. 2017;18:903. doi:10.1093/bib/bbx023
14. Chan J, Wang X, Turner JA, Baldwin NE, Gu J. Breaking the paradigm: Dr Insight empowers signature-free, enhanced drug repurposing. Bioinformatics. 2019;35:2818–2826. doi:10.1093/bioinformatics/btz006
15. Mejia-Pedroza RA, Espinal-Enriquez J, Hernandez-Lemus E. Pathway-based drug repositioning for breast cancer molecular subtypes. Front Pharmacol. 2018;9:905. doi:10.3389/fphar.2018.00905
16. Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10:1209–1210. doi:10.1038/nmeth.2689
17. Ding S, Chen X, Shen K. Single-cell RNA sequencing in breast cancer: understanding tumor heterogeneity and paving roads to individualized therapy. Cancer Commun. 2020;40:329–344. doi:10.1002/cac2.12078
18. Yuan X, Wang J, Huang Y, Shangguan D, Zhang P. Single-cell profiling to explore immunological heterogeneity of tumor microenvironment in breast cancer. Front Immunol. 2021;12:643692. doi:10.3389/fimmu.2021.643692
19. He B, Xiao Y, Liang H, et al. ASGARD is a single-cell guided pipeline to aid repurposing of drugs. Nat Commun. 2023;14:993. doi:10.1038/s41467-023-36637-3
20. Greene CS, Voight BF. Pathway and network-based strategies to translate genetic discoveries into effective therapies. Hum Mol Genet. 2016;25:R94–R8. doi:10.1093/hmg/ddw160
21. Ma J, Wang J, Ghoraie LS, Men X, Haibe-Kains B, Dai P. A comparative study of cluster detection algorithms in protein-protein interaction for drug target discovery and drug repurposing. Front Pharmacol. 2019;10:109. doi:10.3389/fphar.2019.00109
22. Turanli B, Karagoz K, Bidkhori G, et al. Multi-omic data interpretation to repurpose subtype specific drug candidates for breast cancer. Front Genet. 2019;10:420. doi:10.3389/fgene.2019.00420
23. Duan Q, Reid SP, Clark NR, et al. L1000CDS(2): LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl. 2016;2:16015. doi:10.1038/npjsba.2016.15
24. Peyvandipour A, Saberian N, Shafi A, Donato M, Draghici S. A novel computational approach for drug repurposing using systems biology. Bioinformatics. 2018;34:2817–2825. doi:10.1093/bioinformatics/bty133
25. Firoozbakht F, Rezaeian I, Rueda L, Ngom A. Computationally repurposing drugs for breast cancer subtypes using a network-based approach. BMC Bioinform. 2022;23:143. doi:10.1186/s12859-022-04662-6
26. Islam MM, Wang Y, Hu P. A maximum flow-based approach to prioritize drugs for drug repurposing of chronic diseases. Life. 2021;11:54.
27. Saha Detroja T, Detroja R, Mukherjee S, Samson AO. Identifying hub genes associated with neoadjuvant chemotherapy resistance in breast cancer and potential drug repurposing for the development of precision medicine. Int J Mol Sci. 2022;23:12628. doi:10.3390/ijms232012628
28. Di J, Zheng B, Kong Q, et al. Prioritization of candidate cancer drugs based on a drug functional similarity network constructed by integrating pathway activities and drug activities. Mol Oncol. 2019;13:2259–2277. doi:10.1002/1878-0261.12564
29. Al-Taie Z, Hannink M, Mitchem J, Papageorgiou C, Shyu CR. Drug repositioning and subgroup discovery for precision medicine implementation in triple negative breast cancer. Cancers. 2021;13:6278. doi:10.3390/cancers13246278
30. Hsu JL, Hung MC. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 2016;35:575–588. doi:10.1007/s10555-016-9649-6
31. Balbuena-Rebolledo I, Padilla M, Rosales-Hernandez MC, Bello M. Repurposing FDA drug compounds against breast cancer by targeting EGFR/HER2. Pharmaceuticals. 2021;14:791. doi:10.3390/ph14080791
32. Kandasamy T, Sen P, Ghosh SS. Multi-targeted drug repurposing approach for breast cancer via integrated functional network analysis. Mol Inform. 2022;41:e2100300. doi:10.1002/minf.202100300
33. Olsauskas-Kuprys R, Zlobin A, Osipo C. Gamma secretase inhibitors of Notch signaling. Onco Targets Ther. 2013;6:943–955. doi:10.2147/OTT.S33766
34. Pathak Y, Camps I, Mishra A, Tripathi V. Targeting notch signaling pathway in breast cancer stem cells through drug repurposing approach. Mol Divers. 2022. doi:10.1007/s11030-022-10561-y
35. Rui M, Cai M, Zhou Y, et al. Identification of potential RBPJ-specific inhibitors for blocking notch signaling in breast cancer using a drug repurposing strategy. Pharmaceuticals. 2022;15:556. doi:10.3390/ph15050556
36. Maruca A, Rocca R, Catalano R, et al. Natural products extracted from fungal species as new potential anti-cancer drugs: a structure-based drug repurposing approach targeting HDAC7. Molecules. 2020;25:5524. doi:10.3390/molecules25235524
37. Issa NT, Stathias V, Schurer S, Dakshanamurthy S. Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol. 2021;68:132–142. doi:10.1016/j.semcancer.2019.12.011
38. Song J, Xu Z, Cao L, Wang M, Hou Y, Li K. The discovery of new drug-target interactions for breast cancer treatment. Molecules. 2021;26:7474. doi:10.3390/molecules26247474
39. You J, McLeod RD, Hu P. Predicting drug-target interaction network using deep learning model. Comput Biol Chem. 2019;80:90–101. doi:10.1016/j.compbiolchem.2019.03.016
40. Cui C, Ding X, Wang D, et al. Drug repurposing against breast cancer by integrating drug-exposure expression profiles and drug-drug links based on graph neural network. Bioinformatics. 2021;37:2930–2937. doi:10.1093/bioinformatics/btab191
41. Wang A, Lim H, Cheng SY, Xie L. ANTENNA, a multi-rank, multi-layered recommender system for inferring reliable drug-gene-disease associations: repurposing diazoxide as a targeted anti-cancer therapy. IEEE/ACM Trans Comput Biol Bioinform. 2018;15:1960–1967. doi:10.1109/TCBB.2018.2812189
42. Ji X, Jin C, Dong X, Dixon MS, Williams KP, Zheng W. Literature-wide association studies (LWAS) for a rare disease: drug repurposing for inflammatory breast cancer. Molecules. 2020;25:3933. doi:10.3390/molecules25173933
43. Malik JA, Ahmed S, Momin SS, et al. Drug repurposing: a new hope in drug discovery for prostate cancer. ACS Omega. 2023;8:56–73. doi:10.1021/acsomega.2c05821
44. Luck K, Kim DK, Lambourne L, et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi:10.1038/s41586-020-2188-x
45. Kairys V, Baranauskiene L, Kazlauskiene M, Matulis D, Kazlauskas E. Binding affinity in drug design: experimental and computational techniques. Expert Opin Drug Discov. 2019;14:755–768. doi:10.1080/17460441.2019.1623202
46. Melnekoff DT, Lagana A. Single-cell sequencing technologies in precision oncology. Adv Exp Med Biol. 2022;1361:269–282.
47. Robin V, Bodein A, Scott-Boyer MP, Leclercq M, Perin O, Droit A. Overview of methods for characterization and visualization of a protein-protein interaction network in a multi-omics integration context. Front Mol Biosci. 2022;9:962799.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.