Back to Journals » International Journal of General Medicine » Volume 15
Inducible Co-Stimulator ICOS Expression Correlates with Immune Cell Infiltration and Can Predict Prognosis in Lung Adenocarcinoma
Authors Wu G, He M, Ren K, Ma H, Xue Q
Received 11 January 2022
Accepted for publication 9 March 2022
Published 6 April 2022 Volume 2022:15 Pages 3739—3751
DOI https://doi.org/10.2147/IJGM.S349441
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Gujie Wu,1,* Min He,1,* Kuan Ren,1,* Huiyun Ma,1 Qun Xue2
1Medical School of Nantong University, Nantong, People’s Republic of China; 2Cardiothoracic Surgery Department, Affiliated Hospital of Nantong University, Nantong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qun Xue, Email [email protected]
Background: Inducible co-stimulator (ICOS) is a cell-enhanced co-stimulatory receptor that has shown great potential in the regulation of innate and adaptive immunity. However, the role of ICOS in lung adenocarcinoma (LUAD) remains unclear.
Methods: We used data from the Cancer Genome Atlas(TCGA) database to identify the expression and prognostic role of ICOS in LUAD. The results were validated using Gene Expression Omnibus(GEO) and Kaplan-Meier plotter databases. A model with predictive performance for overall survival of LUAD patients was constructed using fitted ICOS expression and other clinical parameters. We explored the biological function of ICOS. Subsequently, we further analysed and validated the effect of ICOS expression on tumour immune microenvironment (TIME) and survival. Finally, the CellMiner database was used to determine the relationship between ICOS expression and drug sensitivity.
Results: ICOS expression is significantly associated with poor prognosis in multiple cancers, especially LUAD, and is a good predictor of overall survival in LUAD patients. The biological function is to promote autoimmunity and inhibit cell proliferation. ICOS-related survival prediction model developed to more accurately predict 1-, 3- and 5-year survival probabilities for LUAD patients. In addition, we can use the expression of ICOS to effectively assess patient malignancy, prognosis, TIME status and clinical combination of drugs.
Conclusion: Our results suggest that ICOS is correlated with prognosis and immune infiltrating levels in LUAD. Higher ICOS expression predicts better TIME. This study provides a novel strategy for the development of immunotherapeutic and prognostic markers in LUAD.
Keywords: ICOS, lung adenocarcinoma, prognosis, immune infiltration, drug sensitivity
Introduction
Lung cancer is one of the most common malignancies, of which more than 80% is non-small cell lung cancer (NSCLC). Lung adenocarcinoma (LUAD), the most recurrent of these NSCLC subtypes, accounts for more than 40% of all lung cancer cases.1,2 Over the past few years, although targeted therapies and immunotherapy have improved the prognosis of LUAD patients have improved clinical outcomes, the 5-year survival rate is still below 20%.3–5 Therefore, finding more accurate markers and therapeutic targets for LUAD is urgently needed, which is the key to improving the survival rate of LUAD patients.
Inducible T cell costimulator (ICOS) is a CD28 superfamily receptor induced in activated T lymphocytes that regulates T cell activation and participates in adaptive immune responses.6 It has been demonstrated that they are aberrantly expressed in a variety of tumors and are associated with cancer development, metastasis and disease.7–10 It has been reported that modulation of ICOS expression may improve anti-tumor immunity, and ICOS co-stimulation at tumor sites combined with anti-CTLA-4 and Treg depletion may also exhibit higher anti-tumor effects.11,12 Although immunotherapy with ICOS is promising, there are still too few studies to date and the mechanism of action is still not well understood. Therefore, the assessment of ICOS as an immune-related biomarker in LUAD and its potential use remains to be further explored.
In this study, we analysed the expression and prognosis of ICOS in pan-cancer and found that ICOS was significantly associated with poor prognosis and poorer clinical factors in several cancers, particularly LUAD. The mechanism of ICOS in the development of LUAD was revealed and analysed in relation to the level of immune cell infiltration and the tumour immune microenvironment. The findings in this study elucidate the critical role of ICOS as a prognostic biomarker in the diagnosis and development of LUAD and reveal potential mechanisms between ICOS and tumour immunity, which provide a basis for clinical research and immune-targeted therapy.
Materials and Methods
Data Collection
The expression profiles and clinical information of TCGA were downloaded from the TCGA database (https://portal.gdc.cancer.gov/).
The expression profiles and clinical information of GSE72094 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds).
Oncomine Database
Oncomine is a database that aggregates multiple cancer types and multiple datasets (https://oncomine.org/resource/login.html).13 Using data from this database, we validated the mRNA expression of ICOS in LUAD.
Kaplan-Meier Plotter Database Analysis
The Kaplan-Meier Plotter can evaluate the impact of 54,675 genes on survival using 10,461 cancer samples. We validated the correlation between ICOS expression and survival in LUAD using data from the Kaplan-Meier Plotter.
Linkedomics Database
LinkedOmics is a database for GO, KEGG and Reactome functional analysis online based on a large number of tumour samples (http://www.linkedomics.org/login.php).
TISIDB Database
TISIDB (http://cis.hku.hk/TISIDB) is a strong website that provides a huge number of tumor immune-related data and allows for a more in-depth analysis of tumor-immune interactions. The site has 988 genes associated to anti-tumor immunity after grouping 4176 entries from 2530 publications. For each of the 30 TCGA cancer types, associations between genes and immunological activities (eg, lymphocytes, immunomodulators, and chemokines) were pre-calculated. We investigate the immune system connection between ICOS and various tumors in this study.
Statistical Analysis
The statistical significance of differential ICOS expression was determined using the Wilcoxon test. Survival curves with HR and P or Cox P values were plotted, and the correlation and statistical significance of ICOS expression with the degree of immune infiltration of the associated cancer was assessed using the Spearman correlation method. The median was taken as the cutoff point. p ≤ 0.05 was considered statistically significant.
Results
The mRNA Expression Levels of ICOS in Different Types of Human Cancers
To assess the relevance of ICOS in tumour development, we analyzed the differences of ICOS expression levels between tumor and adjacent normal tissues in pan-cancer (Figure 1A). We found that ICOS mRNA expression was significantly higher in Breast invasive carcinoma (BRCA), Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), Esophageal carcinoma (ESCA), Head and Neck squamous cell carcinoma (HNSC), Kidney renal clear cell carcinoma (KIRC), Kidney renal papillary cell carcinoma (KIRP), Liver hepatocellular carcinoma (LIHC), Stomach adenocarcinoma (STAD) and Uterine Corpus Endometrial Carcinoma (UCEC). However, ICOS expression was significantly lower in Lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC) and Thyroid carcinoma (THCA) compared to adjacent normal tissues. Analysis based on 103 tumour samples and 113 paraneoplastic samples from the Oncomine database in Figure 1B and C and 535 tumour samples and 59 paraneoplastic samples from the TCGA database in Figure 1D, E showed that ICOS expression was lower in LUAD than in non-cancerous tissues. In addition, immunohistochemical results provided by the HPA database confirmed similar results (Figure 1F and G). Given its prognostic value in LUAD, we used TCGA normal and LUAD data to generate ROC curves to further analyze the diagnostic value of ICOS in LUAD. Figure 1H shows that the area under the curve (AUC) area was 0.726, indicating that ICOS has the diagnostic ability to distinguish LUAD from normal controls. The results of our analysis suggest that ICOS expression in LUAD is significantly weaker than in normal tissue and may play an important regulatory role in the progression of LUAD, promising to be a diagnostic biomarker for LUAD.
Prognostic Value of ICOS in Cancers Prognosis
Our analysis of clinical data showed that high ICOS expression was associated with better survival in several cancer types (Figure 2A), including LUAD, HNSC, Ovarian serous cystadenocarcinoma (OV), Rectum adenocarcinoma (READ), Sarcoma (SARC), Skin Cutaneous Melanoma (SKCM), and UCEC. In contrast, high ICOS expression resulted in poor prognosis in some cancers such as Lower Grade Glioma (LGG) and Uveal Melanoma (UVM). To avoid noncancer events leading to bias, we also performed an analysis of disease-specific survival (DSS) and progression-free interval (PFI), as shown in Figure 2B and C, respectively. The results showed, as did the OS analysis, that higher ICOS expression was significantly associated with better DSS in LUAD, HNSC, OV, SKCM, UCEC, and UCS. In LUAD, Bladder Urothelial Carcinoma (BLCA), CESC, HNSC, and UCEC, higher ICOS expression was significantly associated with better PFI. To further confirm the importance of ICOS in the survival of patients with LUAD, we analyzed the LUAD data using the Kaplan-Meier Plotter database. Figure 2D shows the OS, FPS (first progression survival), and PPS (post progression survival) of patients with LUAD. This analysis is consistent with the TCGA analysis and confirms that ICOS expression affects the prognosis of patients with LUAD.
Correlations Between ICOS Expression and Clinicopathological Parameters in LUAD Patients
We then analyzed the correlation between expression levels and clinical features of ICOS. Our data were derived from gene expression and clinical data of 535 LUAD patients from TCGA (Table 1). We divided LUAD patients into two groups of high and low expression based on median values and then performed the Wilcoxon test to assess the significance of ICOS expression in different clinical features (Figure 3A–J). ICOS overexpression correlated significantly with T-phase, TNM-phase, response to immunotherapy, gender, and age. Following univariate and multivariate analyses of clinical features, we found that ICOS expression (HR=0.657, P=0.016), T stage (HR=1.764, P=0.017) and N stage (HR=2.138, P<0.001) were independently associated with overall survival in patients with LUAD (Figure 3K and L). These results suggest that ICOS expression, T stage and N stage may play a role in determining clinical treatment.
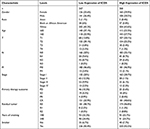 |
Table 1 Grouping of Clinicopathological Features According to ICOS Expression Levels |
Development of a Prognostic Model Based on ICOS and Clinical Factors
An overall survival prediction model was constructed using ICOS and independent clinical risk factors to predict the prognosis of ICOS patients (Figure 4A). In this prediction model, we evaluated these variables and used the summed score of the values of these variables to predict the 1-, 3-, and 5-year survival probabilities of LUAD patients (Figure 4B). The analysis of 1000 bootstrap samples showed that the C-index of ICOS was 0.660 (0.638–0.683). This indicates that the model is highly accurate in terms of predictive performance. Poorer T-stage, N-stage and ICOS expression predicted higher risk scores and poorer prognosis for patients. Thus, the results of our analysis found that the nomogram was a model that was better than individual prognostic factors in predicting short- or long-term survival in LUAD patients.
ICOS Co‐expression Network in LUAD
To understand the biological function of ICOS in LUAD, we used LinkedOmics database analysis to find 10,765 genes (dark red dots) positively linked to ICOS and 9222 genes (dark green dots) negatively linked in TCGA-LUAD (Figure 5A, Supplementary Table 1). Figure 5B and C shows heat maps for the top 50 genes positively and negatively linked to ICOS.GO term annotation showed that co‐expressed genes of ICOS co-expressed genes mainly promote immune processes such as interleukin production, adaptive immune response, interferon-gamma production, T cell activation, leukocyte proliferation, lymphocyte activation involved in immune response, and inhibit cell proliferation processes such as mitochondrial respiratory chain complex assembly and NADH dehydrogenase complex assembly (Figure 5D). Analysis of the KEGG pathway has shown that it promotes autoimmune and inflammatory responses and inhibits oncogenic-related pathways (Figure 5E). Reactome analysis similarly confirmed that ICOS expression promotes autoimmune responses and suppresses tumor cell proliferation during cancer development, we also found that ICOS expression was positively correlated with some immunity-related signalling, such as PD-1 signalling and TCR signalling (Figure 5F). These results suggest that the expression network of ICOS could influence prognosis in LUAD by regulating tumor cell proliferation and autoimmunity.
Immune Infiltration Analysis of ICOS in LUAD
Tumor infiltrating lymphocytes (TILs) have been shown to be an independent predictor of sentinel lymph node (SLN) status and survival.14 To investigate the role of ICOS in the tumor immune microenvironment, we performed correlation analysis using the ESTIMATE R package. We found that ICOS correlated positively with stromal score, ESTIMATE score, and immune score and correlated negatively with tumor clearance in LUAD, and not only that, increased ICOS expression enhances immune cell infiltration and immune function (Figure 6A and B). We next analyzed the relationship between ICOS and 24 types of immune cell infiltration in LUAD (Figure 6C). The results revealed that there was a significant positive correlation between ICOS and almost all immune cell infiltrates, including T cell expression (R=0.810, P<0.001) (Figure 6D). With these results we found that differences in ICOS expression affect the degree of infiltration of different immune cells in LUAD. Finally, to determine the correlation between ICOS expression, TIME and survival of LUAD patients, we performed analysis and validation using data from TCGA and GSE72094, as shown in Figure 6E, in the TCGA (R=0.82, P<2.2e-16) and GSE72094 (R=0.8, P<2.2e-16) datasets. There was a significant positive correlation between ICOS expression and immune score, and patients with high immune score had better overall survival than those with low immune score (Figure 6F). These results suggest that high ICOS expression may contribute to immune cell infiltration and improve the TIME in patients with lung adenocarcinoma, resulting in a better prognosis for patients with LUAD.
Relation Between ICOS with Immune Molecules
We expanded our understanding of the relationship between ICOS and the immune microenvironment by analyzing data from the TISIDB database. We found a particularly strong correlation between ICOS expression and several immunological traits. In most cancers, high ICOS expression can lead to a significant increase in tumor-infiltrating lymphocytes (TILs), and Figure 7A shows the six TILs with the highest correlation, including Tem_CD8 (R=0.788, P<2.2e-16). Figure 7B shows a strong positive correlation between ICOS and immunoinhibitors. Figure 7C shows a correlation between ICOS expression and immunostimulants, with higher ICOS expression stimulating a stronger immune response. Figure 7D shows the correlation between ICOS expression and MHC molecules and demonstrates that high ICOS expression induced a strong immune response to MHC in various tumors. Figure 7E shows the correlation between ICOS expression and chemokines, with the highest correlation between ICOS and CCL5 (R=0.731). Figure 7F shows the correlation between ICOS expression and receptors, with ICOS not only correlating with CXCR6 and CCR5 above 0.80, but also showing strong facilitation with other receptors.
Thus, our study suggests that ICOS as a novel prognostic marker is actively involved in the regulation of various immune molecules in LUAD and influences immune infiltration in the tumor microenvironment.
Gene Mutation Between High and Low-ICOS Groups in LUAD
Gene mutations can promote cancer development and drive malignant cancer proliferation. Investigating mutations at the genomic level is important for the development of tumour-targeted drugs and novel oncology therapies. We divided LUAD patients into two groups according to their median expression of ICOS and compared the differences of gene mutations between the two groups. The results indicated that patients with low ICOS expression possess higher gene mutation frequency than those with high ICOS expression in LUAD (Figure 8). This suggested that low ICOS expression could result in a worse tumour microenvironment and progression in LUAD patients.
 |
Figure 8 Gene mutation between high and low-ICOS groups in LUAD. Top 20 genes with the highest mutation differences between the high-low ICOS groups in LUAD. |
Correlation Between ICOS and Drug Sensitivity
Finally, we investigated the antitumor drugs sensitive to ICOS expression using the CellMiner database. Of these, the sensitivity of 32 drugs was significantly correlated with ICOS expression and the results are shown in Figure 9 and Table 2. ICOS expression was significantly positively correlated with the sensitivity of Pipobroman, Bendamustine, Sapacitabine, Nelarabine and Dabrafenib, and negatively correlated with the sensitivity of Cordycepin and Saracatinib.
 |
Table 2 Correlation of ICOS Expression with the Sensitivity of Anticancer Drugs |
 |
Figure 9 Correlation of ICOS expression with the sensitivity of anticancer drugs. |
Discussion
LUAD is one of the most common types of cancer and a worldwide killer of health.15 In the absence of early symptoms, patients often miss the opportunity to receive the most appropriate treatment.16 Due to the poor prognosis of advanced LUAD, there is an urgent need to find more accurate potential biomarkers to predict the prognosis of patients with LUAD and to personalize their treatment.
ICOS belongs to a family of co-stimulatory factors that are closely associated with tumor development and play an important role in the activation and proliferation of T cells.17,18 Elevated ICOS has been shown to induce its own anti-tumor response not only by regulating immune cell homeostasis but also by regulating the production of inflammatory cytokines,19–21 such as TNF-α and IFN-γ, which contribute to T cell immune function.22–24 In primary breast cancer, ICOS mediates the interaction between tumor-infiltrating CD4+ T cells and plasmacytoid dendritic cells (pDCs), leading to the proliferation of regulatory T cells (Tregs) and the secretion of interleukin 10, which alters the immune microenvironment of the tumor and thus affects patient survival.25,26 In addition, ICOS expression in cholangiocarcinoma and metastatic melanoma has been shown to significantly improve survival by modulating autoimmunity.17,27 Not only can ICOS inhibit tumor progression and enhance immune cell activity, but the combination of anti-PD-L1 and anti-CTLA-4 therapy can further improve the immune environment of tumors and significantly enhance the anti-tumor immune response.28–30 Therefore, ICOS can be used as a potential prognostic biomarker and target for immunotherapy in the diagnosis and treatment of patients with LUAD.
In this study, in order to investigate the role of ICOS in the prognosis and immunological aspects of LUAD, we collected data from a large number of LUAD patients. Our analysis showed that ICOS expression is not only involved in the pathological tissue progression of LUAD but is also an independent prognostic factor for patients with LUAD. The ICOS prognostic model showed good agreement between actual and predicted survival at 1, 3, and 5 years in LUAD patients and may be a valuable new prognostic approach for future clinicians. ICOS improves the prognosis of LUAD primarily by inhibiting tumor progression and modulating autoimmune-related signaling pathways. To investigate the Immunological mechanisms, we found that high expression of ICOS induces increased infiltration of most immune cells, especially T cells. In addition, ICOS is also actively involved in the regulation of various immune molecules. Our results suggest that ICOS can be involved in regulating the immune microenvironment and improving the prognosis of LUAD. Finally, we obtained antitumor agents sensitive to ICOS expression.
In conclusion, our research shows that ICOS is a potential biomarker and therapeutic target associated with immunity and prognosis in patients with LUAD. Low expression of ICOS predicts worse clinical characteristics, TIME and prognosis in patients with LUAD. This study provides new insights into the clinical management of patients with LUAD.
Data Sharing Statement
The datasets generated during the current study are available in the TCGA database (https://portal.gdc.cancer.gov/) and GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Ethics Approval and Consent to Participate
The Ethics Committee of Affiliated Hospital of Nantong University has granted exemptions from approval for research related to the use of such public databases.
Consent for Publication
All authors contributed to the article and approved the submitted version.
Funding
Not applicable.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi:10.3322/caac.21387
2. Shi J, Hua X, Zhu B, et al. Somatic genomics and clinical features of lung adenocarcinoma: a retrospective study. PLoS Med. 2016;13:e1002162. doi:10.1371/journal.pmed.1002162
3. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
4. Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21:2213–2220. doi:10.1158/1078-0432.CCR-14-2748
5. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):
6. Soldevilla MM, Villanueva H, Meraviglia-Crivelli D, et al. ICOS costimulation at the tumor site in combination with CTLA-4 blockade therapy elicits strong tumor immunity. Mol Ther. 2019;27(11):1878–1891. doi:10.1016/j.ymthe.2019.07.013
7. Losurdo A, Scirgolea C, Alvisi G, et al. Single-cell profiling defines the prognostic benefit of CD39 tissue resident memory CD8+ T cells in luminal-like breast cancer. Commun Biol. 2021;4:1117. doi:10.1038/s42003-021-02595-z
8. Fernando C, Behnaz B, Shroff Rachna T, et al. The immunogenomic landscape of resected intrahepatic cholangiocarcinoma. Hepatology. 2021;75:297–308.
9. Sarker MMK, Makhlouf Y, Craig Stephanie G, et al. A means of assessing deep learning-based detection of ICOS protein expression in colon cancer. Cancers. 2021;13. doi:10.3390/cancers13153825
10. Li C, Liu T, Liu Y, et al. Prognostic value of tumour microenvironment-related genes by TCGA database in rectal cancer. J Cell Mol Med. 2021;25:5811–5822. doi:10.1111/jcmm.16547
11. Metzger Todd C, Hua L, Potluri S, et al. ICOS promotes the function of CD4+ effector T cells during anti-OX40-mediated tumor rejection. Cancer Res. 2016;76:3684–3689. doi:10.1158/0008-5472.CAN-15-3412
12. Shi LZ, Goswami S, Fu T, et al. Blockade of CTLA-4 and PD-1 enhances adoptive T-cell therapy efficacy in an ICOS-mediated manner. Cancer Immunol Res. 2019;7:1803–1812. doi:10.1158/2326-6066.CIR-18-0873
13. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi:10.1593/neo.07112
14. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi:10.1158/0008-5472.can-17-0307
15. Ettinger DS, Akerley W, Bepler G, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8:740–801. doi:10.6004/jnccn.2010.0056
16. Wu CF, Fu JY, Yeh CJ, et al. Recurrence risk factors analysis for stage I non-small cell lung cancer. Medicine. 2015;94:e1337. doi:10.1097/MD.0000000000001337
17. He M, Wang Y, Shi WJ, et al. Immunomodulation of inducible co-stimulator (ICOS) in human cytokine-induced killer cells against cholangiocarcinoma through ICOS/ICOS ligand interaction. J Dig Dis. 2011;12:393–400. doi:10.1111/j.1751-2980.2011.00527.x
18. Amatore F, Gorvel L, Olive D. Inducible co-stimulator (ICOS) as a potential therapeutic target for anti-cancer therapy. Expert Opin Ther Targets. 2018;22:343–351. doi:10.1080/14728222.2018.1444753
19. Burmeister Y, Lischke T, Dahler AC, et al. ICOS controls the pool size of effector-memory and regulatory T cells. J Immunol. 2008;180:774–782. doi:10.4049/jimmunol.180.2.774
20. Wikenheiser DJ, Stumhofer JS. ICOS co-stimulation: friend or foe? Front Immunol. 2016;7:304
21. leconte J, Bagherzadeh Yazdchi S, Panneton V, Suh WK. Inducible costimulator (ICOS) potentiates TCR-induced calcium flux by augmenting PLCγ1 activation and actin remodeling. Mol Immunol. 2016;79:38–46. doi:10.1016/j.molimm.2016.09.022
22. Yoshinaga SK, Zhang M, Pistillo J, et al. Characterization of a new human B7-related protein: B7RP-1 is the ligand to the co-stimulatory protein ICOS. Int Immunol. 2000;12:1439–1447. doi:10.1093/intimm/12.10.1439
23. Kopf M, Coyle AJ, Schmitz N, et al. Inducible costimulator protein (ICOS) controls T helper cell subset polarization after virus and parasite infection. J Exp Med. 2000;192:53–61. doi:10.1084/jem.192.1.53
24. Akbari O, Freeman GJ, Meyer EH, et al. Antigen-specific regulatory T cells develop via the ICOS-ICOS-ligand pathway and inhibit allergen-induced airway hyperreactivity. Nat Med. 2002;8:1024–1032. doi:10.1038/nm745
25. Faget J, Bendriss-Vermare N, Gobert M, et al. ICOS-ligand expression on plasmacytoid dendritic cells supports breast cancer progression by promoting the accumulation of immunosuppressive CD4+ T cells. Cancer Res. 2012;72:6130–6141. PMID:23026134. doi:10.1158/0008-5472.CAN-12-2409
26. Faget J, Sisirak V, Blay JY, Caux C, Bendriss-Vermare N, Menetrier-Caux C. ICOS is associated with poor prognosis in breast cancer as it promotes the amplification of immunosuppressive CD4 T cells by plasmacytoid dendritic cells. Oncoimmunology. 2013;2:e23185. doi:10.4161/onci.23185
27. Carthon Bradley C, Wolchok Jedd D, Yuan J, et al. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin Cancer Res. 2010;16:2861–2871. doi:10.1158/1078-0432.CCR-10-0569
28. Shi LZ, Fu T, Guan B, et al. Interdependent IL-7 and IFN-gamma signalling in T-cell controls tumour eradication by combined alpha-CTLA-4+alpha-PD-1 therapy. Nat Commun. 2016;7:12335. doi:10.1038/ncomms12335
29. Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. PNAS. 2010;107:4275–4280. doi:10.1073/pnas.0915174107
30. Sainson Richard CA, Thotakura Anil K, Kosmac M, et al. An antibody targeting ICOS increases intratumoral cytotoxic to regulatory T-cell ratio and induces tumor regression. Cancer Immunol Res. 2020;8:1568–1582. doi:10.1158/2326-6066.CIR-20-0034
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







