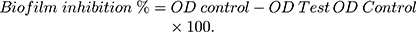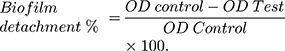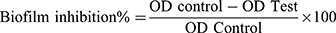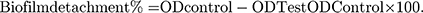Back to Journals » Infection and Drug Resistance » Volume 14
In vivo Antibacterial Activity of Star Anise (Illicium verum Hook.) Extract Using Murine MRSA Skin Infection Model in Relation to Its Metabolite Profile
Authors Salem MA, El-Shiekh RA , Hashem RA, Hassan M
Received 9 October 2020
Accepted for publication 12 December 2020
Published 6 January 2021 Volume 2021:14 Pages 33—48
DOI https://doi.org/10.2147/IDR.S285940
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Mohamed A Salem,1,* Riham A El-Shiekh,2,* Rasha A Hashem,3 Mariam Hassan3
1Department of Pharmacognosy, Faculty of Pharmacy, Menoufia University, Menoufia, Egypt; 2Department of Pharmacognosy, Faculty of Pharmacy, Cairo University, Cairo, Egypt; 3Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo, Egypt
*These authors contributed equally to this work
Correspondence: Mariam Hassan
Faculty of Pharmacy, Cairo University, Kasr El-Aini Street, Cairo 11562, Egypt
Tel +20 122 3376326
Email [email protected]
Introduction: Star anise fruits (Illicium verum Hook.) have been used as an important treatment in traditional Chinese medicine. The previous studies reported the activity of the non-polar fractions as potential sources of antibacterial metabolites, and little was done concerning the polar fractions of star anise.
Methods: The antibacterial activity of the star anise aqueous methanolic (50%) extract against multidrug-resistant Acinetobacter baumannii AB5057 and methicillin-resistant Staphylococcus aureus (MRSA USA300) was investigated in vitro (disc diffusion assay, minimum bactericidal concentration determination, anti-biofilm activity and biofilm detachment activity). The antibacterial activity was further tested in vivo using a murine model of MRSA skin infection. Ultra-performance liquid chromatography coupled to high-resolution mass spectrometry (UPLC/HRMS) approach was applied for the identification of the metabolites responsible for the antibacterial activity. The antioxidant potential was evaluated using five in vitro assays: TAC (total antioxidant capacity), DPPH, ABTS, FRAP (ferric reducing antioxidant power) and iron-reducing power.
Results: In vitro, star anise aqueous methanolic extract showed significant inhibition and detachment activity against biofilm formation by the multidrug-resistant and highly virulent Acinetobacter baumannii AB5057 and MRSA USA300. The topical application of the extract in vivo significantly reduced the bacterial load in MRSA-infected skin lesions. The extract showed strong antioxidant activity using five different complementary methods. More than seventy metabolites from different classes were identified: phenolic acids, phenylpropanoids, sesquiterpenes, tannins, lignans and flavonoids.
Conclusion: This study proposes the potential use of star anise polar fraction in anti-virulence strategies against persistent infections and for the treatment of staphylococcal skin infections as a topical antimicrobial agent. To our knowledge, our research is the first to provide the complete polar metabolome list of star anise in an approach to understand the relationship between the chemistry of these metabolites and the proposed antibacterial activity.
Keywords: Acinetobacter baumannii, antimicrobial resistance, antioxidant, biofilm, metabolome, polar methanolic extract
Introduction
Illicium verum Hook. (F. Schisandraceae) or Chinese star anise is an evergreen tree native to southern China and northern Vietnam and listed in the Chinese Pharmacopoeia (2010 edition).1 The fruit is red to brown, star-shaped, consists of about 8–13 centrally joined carpels.1 It is a well-known spice that closely resembles anise in flavour. Chinese star anise is used in traditional medicine for the treatment of rheumatic pain, stomach aches, skin inflammation, vomiting and insomnia. It also was reported to have anti-flu, anti-HIV (human immunodeficiency virus), antifungal, antiseptic, insecticidal and chemopreventive activities.1 Phenylpropanoids, sesquilignans, shikimic acid, flavonoids, and seco-prezizaane-type sesquiterpenoids are the common classes of compounds previously identified in the plant.1
Acinetobacter baumannii and Staphylococcus aureus belong to the clinically important ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) that are the leading cause of worldwide nosocomial infections.2 A. baumannii was associated with high morbidity and mortality rates including wound infections, blood stream infections, and secondary meningitis, pneumonia and urinary tract infections.3 S. aureus is the most frequent pathogen isolated from human skin and wound infections. One of the greatest challenges in clinical practice is that most of these pathogens are multidrug-resistant (MDR). For instance, methicillin-resistant Staphylococcus aureus (MRSA) is a community and hospital-acquired pathogen that can induce chronic infectious diseases with high morbidity and mortality rates.2,4 Biofilm is a form of bacterial growth unanimously in environmental niches. Its formation leads to a highly raised pattern of adaptive resistance to antibiotics and antimicrobial agents.5 This adaptive resistance acts as a big problem that worsens the infectious diseases as ventilator-associated pneumonia, nosocomial pneumonia, surgical wound, burn wound and catheter-associated infections.6,7 The newly discovered natural antimicrobial and anti-biofilm agents are promising candidates that could provide a novel strategy for fighting those infections.8,9
In this research, we investigated the efficacy of the star anise aqueous methanolic (50%) extract as an anti-biofilm agent in vitro and in vivo using a murine model of MRSA skin infection. Moreover, we identified the bioactive metabolites in the extract using a comprehensive UPLC/HRMS metabolomics approach.
Materials and Methods
Chemicals, Reagents and Instruments
Methyl tert-butyl ether (MTBE), methanol HPLC grade, 2,2ʹ-azino-bis-3-ethylbenzthiazoline-6-sulphonic acid (ABTS), 2,4,6-Tris(2-pyridyl)-1,3,5-triazine (TPTZ), 2, 2-diphenyl-1-picryl-hydrazyl (DPPH), Folin-Ciocalteau, gallic acid, quercetin, 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (trolox), aluminum chloride hexahydrate, dimethylsulfoxide (DMSO) and trichloroacetic acid were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO). Sodium phosphate, hydrochloric acid, ammonium molybdate, sodium carbonate, sodium acetate, potassium persulfate, potassium ferricyanide and ferric chloride were purchased from El-Nasr Company for Pharmaceutical Chemicals, Egypt. Phosphate buffer and Tris-HCl buffer were purchased from Bio diagnostic, Egypt. Waters Acquity UPLC system (Waters, Manchester, UK) hyphenated with Orbitrap-type, HRMS (Exactive, Thermo-Fisher, Bremen, Germany) was used for metabolite profiling.
Plant Materials and Extraction
The fruits of star anise (Illicium verum Hook.) were purchased from Herbal store, Egypt and authenticated by Mrs. Teresa Labib, the senior taxonomist at Orman Botanic Garden. A voucher specimen (No. 15.7.2019) was placed in the Herbarium of the Department of Pharmacognosy, Faculty of Pharmacy, Cairo University, Egypt. The dried powder (1 kg) was extracted with MTBE: methanol (3:1 v/v, 3x2500 mL) followed by water-methanol (3:1 v/v, 3x2500 mL) by adopting the method of Salem et al.10 After that, the extract was fractionated, evaporated under reduced pressure till dryness and then kept in the desiccator over anhydrous CaCl2 till use. Different concentrations (mg/mL) of the dried extract were prepared in analytical grade dimethylsulfoxide (DMSO) to be used in the biological study of the polar fraction (aqueous methanolic extract).
In all the in vitro experiments, the extract was prepared in DMSO to the desired concentrations, and so DMSO was used as the negative control in all the in vitro experiments. In the in vivo model, a stock solution (400 mg/mL) of the plant extract was prepared in DMSO then diluted with water (25%v/v) to get the desired tested concentration (100 mg/mL); hence, the negative control (vehicle control) was 25%v/v DMSO in water.
Bacterial Strains
The highly virulent Acinetobacter baumannii AB505711 and methicillin-resistant Staphylococcus aureus (MRSA USA300)12 were used as the test organisms in this study.
Screening of Antibacterial Activity
The standard Kirby–Bauer procedure (disc diffusion method) was used to evaluate the antibacterial activity of the extract.13 Mueller Hinton agar plates were surface inoculated (swabbed) with active bacterial cultures (pre-adjusted at OD600 to 0.1±0.02, 0.5 McFarland standard suspensions). Sterile filter paper discs (Whatman no. 5, 6 mm in diameter) were loaded with the tested extract at a concentration of 10 mg/disc; the discs were then placed on the inoculated agar surface. The plates were incubated at 37°C overnight (18 h) under aerobic conditions. After the incubation period, the diameters of the zone of inhibition were measured. Dimethylsulfoxide (DMSO) was used as a negative control. Tetracycline (30 μg/disc) and vancomycin (5 μg/disc) were used as positive controls. All the experiments were repeated three independent times.
Minimum Bactericidal Concentration (MBC) Assay
The minimum bactericidal concentration (MBC) determination was conducted using the broth microdilution method,13 where 100 μL of double strength Mueller Hinton broth was placed in each well of a sterile 96-well plate and 100 μL of the tested extract (100mg/mL) or DMSO (negative control) was then added to the first well of each row. Two-fold serial dilutions were done from one row to the next till the eighth row (50–0.391 mg/mL). The wells were then inoculated with 10 μL of the tested bacterial suspension (108 CFU/mL). One row was used as a sterility control (neither bacterial suspension nor tested extract was added) and another row was used as a growth control (inoculated with bacterial suspension without adding the tested extract). Plates were incubated at 37°C for 24 h after which 5 μL of each well was spotted on Mueller Hinton agar plate then incubated at 37°C for 24 h. MBC was determined as the lowest concentration with no detectable colonies. The experiment was repeated three independent times.
Anti-Biofilm Activity (Crystal-Violet Biofilm Formation Assay)
A static biofilm formation assay was performed as described before.11 Briefly, the bacterial suspension in lysogeny broth (LB) broth for A. baumannii AB5057 and tryptic soy broth (TSB) for MRSA USA300 (108 CFU/mL) was loaded in a flat-bottom 96-well ELISA plate (120 μL/well). The bacterial suspension was supplemented with different concentrations of the tested extract (12 μL/well). The tested extract was tested at concentrations below the MBC (0.625, 0.313 and 0.156 mg/mL). For the control wells (untreated wells, 100% reference values), DMSO was added to the bacterial suspension (12 μL/well). The plates were incubated at 37°C for 24 h at static conditions. After the incubation period, optical density (OD600) of the grown cultures was measured by an automated spectrophotometric plate reader (Biotek, Synergy 2, USA). Wells were washed three times with saline then dried thoroughly. The dry biofilm was stained with crystal violet (0.1% w/v, 150 μL/well) for 30 min at room temperature. The wells were then washed three times with distilled water and then dried thoroughly. The crystal violet in the biofilm was solubilized by adding absolute ethanol (150 μL/well) and incubating for 20 min at 4°C. The OD550 of crystal violet solutions was measured by the automated spectrophotometric plate reader, and divided by OD600 of the grown cultures for normalization. The experiment was repeated three independent times. The biofilm inhibition % was calculated using the following equation:
Biofilm Detachment Assay
The biofilm detachment assay was used to test the activity of the tested extract against the previously established biofilm. The assay was performed as described before.14 The bacterial suspension in LB/TSB broth (106 CFU/mL) for A. baumannii AB5057 and MRSA USA300, respectively, was loaded in a flat-bottom 96-well ELISA plate (120 μL/well) and incubated at 37°C for 24 h in static conditions. After the incubation period, OD600 of the grown cultures was measured by an automated spectrophotometric plate reader (Biotek, Synergy 2, USA). The wells were emptied by aspiration. Different concentrations of the tested extract were prepared in fresh broth and were added to the biofilm plate (120 μL/well). Untreated biofilm (control, 100% reference value) was obtained by using DMSO instead of the tested extract. The plate was then incubated at 37°C for 24 h. After the treatment period, crystal violet staining and measurement were done as described above. The experiment was repeated three independent times. The biofilm detachment % was calculated using the following equation:
Efficacy of Star Anise Extract in an in vivo Murine Model of MRSA Skin Infection
The murine model of MRSA skin infection was approved by the Research Ethics Committee of the Faculty of Pharmacy, Cairo University (Approval no. MIC2669) in accordance with the “Guide for the Care and Use of Laboratory Animals” published by the Institute of Laboratory Animal Research (Washington, DC, USA). The murine skin infection model was conducted as described before.15 Eighteen female BALB/C mice (8 weeks old) were obtained from the Modern Veterinary Office for Laboratory Animals, Cairo, Egypt. Mice were allowed to acclimate for one week (25 ± 2 OC, 12:12 h light-dark regime) before starting the experiment. Animals were supplied with the standard commercial food and tap water ad libitum. The posterior upper backs of the mice were shaved one day before starting the experiment. The mice were injected intradermal with 40 µL of mid logarithmically grown S. aureus USA300 suspended in sterile saline (5.5 × 108 CFU). Mice were distributed randomly into three groups; each group contained six animals (n=6). Forty-eight hours post-infection, the first group was topically treated with star anise extract at a concentration of 4MBC (100mg/mL). The second group was used as vehicle control and was topically treated with the vehicle (25%v/v DMSO in water). The third group did not receive any treatment and was used as the negative control. The respective groups were treated once daily for three days. The topical application was done by applying 100 μL of the corresponding treatment at the site of infection. Mice were euthanized after 24 hours from the last treatment. The skin lesion was excised and homogenized in 0.5 mL saline (homogenizer, DAIHAN-scientific-pacificlab). Samples were 10 folds diluted and plated for aerobic viable count on mannitol salt agar (MSA) then incubated at 37°C for 24 hours. The colony-forming units (CFU) were counted and the results of the three groups were compared.
Analysis of Metabolites by UPLC/HRMS
The aqueous methanolic extract (2 µL) was injected and separated on a reversed-phase (RP) C18 column using a UPLC system 10. The mass spectra were acquired in positive and negative ionization modes using a heated electrospray ionization (HESI) source in combination with an Exactive, Orbitrap-type HRMS 10. Metabolites were identified by their mass spectra and comparison with the in house database, plant dictionary database and the references literature.
Phytochemical Assessment of the Total Phenolics and Total Antioxidant Activities
The total phenolic and total flavonoid contents were determined using the method adopted by Aryal et al (2019).16 Briefly, fifty μL of the tested extract (1 mg/mL) or standard solution of gallic acid (6.25–100 μg/mL) were separately added to 50 μL of distilled water, 50 μL of 10% Folin Ciocalteu’s phenol reagent and 50 μL of 1 M sodium carbonate solution in a microplate. Distilled water was used as blank. Plates were incubated for 60 min at room temperature in the dark and absorbance was measured at 750 nm. The total phenolic content was expressed as μg gallic acid equivalents (GAE) per mg of extract.
Total flavonoid content was expressed as μg quercetin equivalents (QE) per mg of plant extract. Briefly, 50 μL of the extract (1 mg/mL) or standard solution of quercetin (6.25–100 μg/mL) were separately added to 10 μL of 10% aluminium chloride solution and 150 μL of 95% ethanol. Ethanol 95% was used as a blank. Plates were incubated for 40 min at room temperature in the dark and absorbance was measured at 415 nm. All samples were analyzed in triplicates. All results were expressed as means of triplicate analyses.
The antioxidant potential was evaluated using five in vitro assays: TAC (total antioxidant capacity), DPPH, ABTS, FRAP (ferric reducing antioxidant power) and iron-reducing power. Sample preparation and analysis were performed as described previously.16 The antioxidant activities were expressed as trolox equivalent (TE) (μmol TE/100 g dry weight).
Total Antioxidant Capacity Assay (TAC)
The antioxidant activity of the samples was tested by the phosphomolybdenum method.16 Where, 250 µg/mL of the extract was prepared in methanol (0.5 mL) and mixed with 3 mL of the following mixture; 28 mM sodium phosphate, 1% ammonium molybdate, and 0.6 M sulphuric acid. After incubation at 95°C for 150 min, the absorbance was measured at 695 nm.
DPPH Radical Scavenging Assay
Different concentrations of the standard (0.1575–1 mg/mL), 250 µg/mL of the extract and DPPH (0.04 g%) were prepared in methanol. Where, 20 μL of each diluted concentrations of the extract or standard were added to 200 μL of DPPH in a 96 well plate, after an incubation period of 30 min in the dark, the absorbance was measured at 492 nm.
ABTS Radical Scavenging Assay
ABTS radical assay was assessed as follows: 10 µL extract at 250 µg/mL or standard at different concentrations (0.1575–1 mg/mL) were separately added to 200 µL ABTS solution and measured at 734 nm in a microplate. The solution of ABTS (5 mL of 14 mM ABTS dissolved in methanol added to 88 μL solution of potassium persulfate 140 mM) was prepared and subsequently stored for 16 hours at room temperature in dark. This was diluted with methanol to reach an absorbance of 0.7.
Ferric Reducing Antioxidant Power (FRAP) Assay
As a FRAP reagent was prepared as follows: 300 mM sodium acetate buffer (pH 3.6, 10 mL),10 mM TPTZ solution in 40 mM hydrochloric acid (1 mL) and 20 mM ferric chloride (1 mL). The FRAP reagent was used in a water bath at 37°C. Then, 10 μL of 250 µg/mL or standard at different concentrations (0.1575–1 mg/mL) was separately mixed with the FRAP reagent (190 μL). The absorbance was determined at 593 nm immediately.
Reducing Power Assay
Different concentrations (0.1575–1 mg/mL) of the standard in methanol or sample at 250 µg/mL (2.5 mL) were added to 2.5 mL of 0.2 M phosphate buffer (pH 6.6) and 2.5 mL of 1% potassium ferricyanide. The mixtures were kept in a water bath at 50°C for 20 minutes. After cooling, 2.5 mL of trichloro acetic acid (10%) was added and centrifuged as required. The prepared solution upper layer was combined with distilled water and 0.1% ferric chloride solution in a ratio of 1:1:2. The absorbance was measured at 700 nm using ascorbic acid as a standard.
Results
Antibacterial Activity and Minimum Bactericidal Concentration (MBC)
Star anise extract was tested for its antimicrobial activity against A. baumannii AB5057 and methicillin-resistant Staphylococcus aureus (MRSA USA300) (Figure S1). The results showed that A. baumannii AB5057 was susceptible to the tested extract and tetracycline with inhibition zone diameter of 13±1 and 25±2 mm, respectively (t-test, P=0.0008). MRSA USA300 showed susceptibility to the tested extract and vancomycin with inhibition zone diameter of 13±0.8 and, 15±0.6 mm, respectively (t-test, P=0.0106). The MBC results confirmed that the tested extract was potent against A. baumannii AB5057 and MRSA USA300 with MBC of 6.25 and 25 mg/mL, respectively.
Anti-Biofilm Activity and Biofilm Detachment Activity
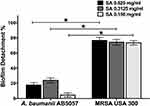
Star anise extract significantly inhibited A. baumannii AB5057 and MRSA USA300 biofilm formation at all the tested concentrations when compared to the control untreated wells (Two-way ANOVA, Tukey’s post-test, P < 0.001) (Figure 1). The tested extract significantly detached the previously formed biofilm by MRSA USA300 at all the tested concentrations when compared to the control untreated wells (Two-way ANOVA, Tukey’s post-test, P < 0.001) (Figure 2). Interestingly, the tested extract showed significantly higher biofilm detachment activity against MRSA USA300 than against A. baumannii AB5057 at all the tested concentrations (Two-way ANOVA, Tukey’s post-test, P < 0.001) (Figure 2).
In vivo Efficacy of Star Anise Extracts Against MRSA Skin Infection in Mice
Three groups of female BALB/C mice (n=6) were injected intradermal with highly virulent MRSA USA300. The mice developed abscess at the site of infection 24 hours post-infection. Treatment with star anise extract significantly reduced the bacterial count of MRSA USA300 compared to negative control and vehicle control groups (One-way ANOVA, Tukey’s post-test, P<0.001) (Figure 3). The bacterial load recovered from the tested group was 3.651 and 3.802 logs lower than that of the vehicle control and negative control groups, respectively. There was no significant difference between the bacterial load recovered from the vehicle control and the negative control groups (One-way ANOVA, Tukey’s post-test, P<0.001).
Analysis of Metabolites by UPLC/HRMS
Star anise metabolites were analyzed by using liquid chromatography coupled with ion spray mass spectrometry in tandem mode (LC/MS/MS) with both positive and negative ion detections (Figures S2A & S2B). Identification of the metabolites was based on their retention times and mass spectra in full scan (MS) mode as well as all-ion-fragmentation (MS/MS) mode. For the first time, a complete list of polar metabolites of star anise was provided (Table 1).
 |
Table 1 Metabolites Identified from the Star Anise Extract After UPLC/MS Analysis |
An exemplary description for the identification of compounds from star anise extract is provided. For instance, the extracted ion chromatograms (EIC) of a protonated compound at m/z 327.14 [M+H]+ and a retention time (Rt) of 9.05 min was detected (Figure 4A). The predicted molecular formula for this adduct was C16H23O7. The MS spectra of the compound showed an ammoniated [M+NH4]+ and sodiated [M+Na]+ adducts that were also detected at m/z 344.17 and 349.13, respectively (Figure 4A). In negative ionization mode, a deprotonated adduct at m/z 325.13 [M-H]− and a deprotonation followed by addition of formic acid [M+FA-H]− at m/z 371.13 were detected at the same Rt (9.05) (Figure 4B). Additionally, highly intense peaks for a protonated fragment after a loss of hexoside moiety [M-glucose+H]+ and a deprotonated fragment of after a loss of hexoside moiety [M-glucose-H]− were detected at m/z 165.09 and 163.08, respectively. These fragments were nicely co-eluted as their precursor adduct at the same Rt (9.05) (Figure 4A and B).
Interestingly, the peak at m/z 165.09117 showed a predicted molecular formula of C10H13O2. Searching the molecular formula to common MS databases such as METLIN (http://metlin.scripps.edu), MassBank (www.massbank.jp), the Human Metabolome Database (http://www.hmdb.ca/), FooDB (https://foodb.ca/) and ChemSpider (http://www.chemspider.com/) showed that the formula could be assigned to a protonated eugenol. This peak, as well as its deprotonated form, was nicely co-eluted as their precursor adduct at the same Rt (9.05) (Figure 4A and B). The molecular formula (C16H22O7) for the compound was tentatively assigned to eugenol glucoside. Comparing the MS and MS/MS spectra of this compound to the available MS data of the FooDB database showed the characteristic pattern of eugenol glucoside and its fragments in positive as well as negative ionization modes (Figures S3A & S3B). Moreover, comparing the MS and MS/MS spectra of the peak at m/z 165.09117 [predicted molecular formula of C10H13O2] to the available MS/MS data of METLIN database showed the pattern of eugenol and its fragments (Figure S4).
Using the same identification protocol, other glycosides of eugenol were also identified. For instance, the EIC of a protonated compound at m/z 473.20 [M+H]+ and a retention time (Rt) of 8.03 and 8.50 min were detected (Figure 5A). The predicted molecular formula for this adduct was C22H33O11. The MS spectra of the compound showed an ammoniated [M+NH4]+ and sodiated [M+Na]+ adducts that were also detected at m/z 490.23 and 495.18, respectively (Figure 5A). In negative ionization mode, a deprotonated adduct at m/z 471.19 [M-H]− and a deprotonation followed by addition of formic acid [M+FA-H]− at m/z 517.19 were detected at the same Rt (8.03 and 8.50 min) (Figure 5B). Additionally, intense peaks for a dimer of ammoniated adduct at m/z 962.42 as well as loss of water molecules from the protonated adduct at m/z 455.19, 437.18 and 419.17 were also detected and were found to co-elute as their precursor adduct at the same Rt (Figure 5A). Intestinally, protonated fragments for loss of deoxy-hexose [M-rhamnose+H]+ and loss of hexoside moiety [M-glucose+H]+ were detected at m/z 327.14 and 311.15, respectively. Searching the predicted molecular formula of the fragment at m/z 165.09117 (C10H13O2) to common MS databases showed the pattern of eugenol (Figure S5). The molecular formula (C22H32O11) for the compound was tentatively assigned to eugenol rutinoside. Comparing the MS and MS/MS spectra of this compound to the available MS data of the METLIN database showed the characteristic pattern of eugenol rutinoside and its fragments in positive as well as negative ionization modes (Figures 4, S5A & S5B).
Similarly, the EIC of a protonated compound at m/z 459.19 [M+H]+ and a retention time (Rt) of 8.35 min was detected (Figure 6A). The predicted molecular formula for this adduct was C21H31O11. The MS spectra of the compound showed an ammoniated [M+NH4]+ and sodiated [M+Na]+ adducts that were also detected at m/z 476.21 and 481.17, respectively (Figure 6A). In negative ionization mode, a deprotonated adduct at m/z 457.17 [M-H]− and a deprotonation followed by addition of formic acid [M+FA-H]− at m/z 503.18 were detected at the same Rt (8.35 and 8.50 min) (Figure 6B). Additionally, intense peaks for a dimer of ammoniated adduct at m/z 934.39 was found to co-elute as the precursor adduct at the same Rt (Figure 6A). Intestinally, protonated fragments for loss of pentose [M-arabinose+H]+ and loss of hexoside moiety [M-glucose+H]+ were detected at m/z 327.14 and 297.13, respectively. Searching the predicted molecular formula of the fragment at m/z 165.09117 (C10H13O2) to common MS databases the pattern of eugenol (Figure S6). The molecular formula (C21H30O11) for the compound was tentatively assigned to eugenol-arabinofuranosyl-glucopyranoside. Comparing the MS and MS/MS spectra of this compound to the available MS data of HMDB showed the characteristic pattern of eugenol-arabinofuranosyl-glucopyranoside and its fragments in positive as well as negative ionization modes (Figures 6, S6A & S6B). Moreover, the chromatographic elution order for these eugenol glycosides was denoted as eugenol rutinoside (8.05 min) < eugenol-arabinofuranosyl-glucopyranoside (8.35 min) < eugenol glucoside (9.05 min), consistent with the elution pattern expected from a reversed-phase column.
Star anise extract exhibited also a mixture containing molecular ions for proanthocyanidin monomers, dimers, trimers and tetramers (Figure S7A & S7B). The EIC of a deprotonated and protonated adducts were detected at m/z 865.19855 and 867.21368, respectively, eluted at retention time (Rt) of 5.20 min (Figure S8). The predicted molecular formula for this compound was C45H37O18 and C45H39O18, respectively. In negative ionization mode, these compounds exhibited the characteristic product ions at m/z 847.19360 [M-H-18(H2O)]−, 713.14923 [M-H-152 (C8H8O3)]−, 577.13519 [M-H-152(C8H8O3)-136(C7H4O3)]−, 407.07669 [M-H-152(C8H8O3)-136(C7H4O3)-152-18(H2O)]−, 289.07150 [M-289(C15H13O6)-288(C15H12O6)]−, 271.08163 [M-289(catechin moiety)-288(C15H12O6)-18(H2O)]− and 245.04517 [M-H-110-165(C8H5O4)-44]−. In addition to these product ions at m/z 579.14990 [M+H-288(C15H12O6)]+, 349.08957 [M+H-110(dihydroxybenzene moiety)-165-44(C2H4O)]+ and 289.07101 [M-288(C15H12O6)]+ were detected in positive ionization mode. Searching the molecular formula and the fragments to common MS databases such as METLIN (http://metlin.scripps.edu), and reviewing literature showed that the formula could be assigned to a protonated proanthocyanidin trimer.17
Star anise is also rich in different classes of compounds including flavonoids. The EIC of a deprotonated and protonated compound at m/z 623.16113 and 625.17712 at retention time (Rt) of 7.82 min was detected (Figure S9). The observation of glycosidic residues (rhamnosyl and glucosyl) was cleaved sequentially and generated characteristic aglycone fragment at m/z 315.05063 and 317.06570 of isorhamnetin. The compound could be identified as isorhamnetin O-rutinoside.
Phytochemical Assessment of the Total Phenolics and Total Antioxidant Activities
Quantitative determinations of the total phenolic and flavonoid contents were estimated as gallic acid equivalent (GAE) and quercetin equivalent (QE), respectively (Table 2). The results revealed that star anise extract was rich in total phenolics and flavonoids contents (309.798 ±6.72 GAE/100 gram dry weight and 98.0±5.79 QE/100 gram dry weight). The in vitro antioxidant activities of the extract showed good reducing power and radical scavenging activities (Table 2).
 |
Table 2 Phenolic Content and Antioxidant Potential of Star Anise Extract |
Discussion
Most bacteria are able to form biofilm on biotic and abiotic surfaces, which is a common cause of persistent infections and resistance to wide spectrum antimicrobial agents 6. Bacterial drug resistance increases to several hundred times in biofilm, which is explained by decreased drug permeability and intracellular survival leading to a persistent infection 6. Hence, new strategies to eradicate biofilm or inhibit biofilm formation are urgently needed. The incidence of antibiotic resistance for A. baumannii infections has increased over the last decade 2. A. baumannii and MRSA have been documented to be responsible for a variety of nosocomial infections due to their ability to form biofilm on abiotic surfaces in the hospitals.2,18 It was reported that they have high levels of resistance to many classes of antimicrobials and therefore, the World Health Organization (WHO) listed them among the top priority pathogens for the development of new antimicrobials.19 A. baumannii AB5057 and MRSA USA300 were selected as the test organisms in this study as they are MDR (multidrug-resistant) and highly virulent strains 11. Biofilm formation participates in staphylococcal resistance and may lead to prolonging of inflammation, and delay the process of wound healing and chronic infections.18 MRSA infections represent a serious challenge due to the emergence of resistance towards numerous antibiotics 15. Since biofilm formation is one of the key players in A. baumannii and MRSA pathogenesis, the ability of star anise extract to inhibit biofilm formation by A. baumannii AB5057 and MRSA USA300 was proved. In addition, the biofilm detachment activity of the star anise extract and the efficacy of it in murine model skin infection were also assessed.
Though the fruits of star anise have been used as a well-known spice and a natural medicine for thousands of years, there has been little research tracing the potent antibiotic and antioxidant activities of star anise with lacking information on its complete chemical profile. Star anise oil has been reported to have antifungal activity owning to its anethole content.20 Additionally, the diethyl ether fraction of I. verum exerted promising antibacterial activity against Acinetobacter baumannii, Pseudomonas aeruginosa, and MRSA.21 Moreover, anethole showed substantial activity against A. baumannii compared to other major constituents such as anisyl alcohol, anisyl aldehyde and anisyl acetone.21 Most of these studies concern the activity of the non-polar compounds of the essential oils (low molecular weight volatile phenolic compounds). These studies have also shown the contribution of the non-polar fraction of star anise as a potential source of antibacterial metabolites 1. Terpenoids or isoprenoids, which are the major part of plant essential oils, represent the most diverse class of biogenic volatile organic compounds in plants that are highly influenced by biotic and environmental factors such as light, temperature, soil water, soil fertility and salinity.22
Therapeutically, star anise has been shown to contain several phenolic compounds 1 however, the antimicrobial activity of its polar metabolites against antibiotic-resistant bacteria has not been deeply investigated, and the whole set of its metabolome has not been identified to date. Therefore, the aim of the present study was to investigate the antibacterial activity of the aqueous methanolic extract from I. verum against the multidrug-resistant and highly virulent A. baumannii AB5057 and MRSA USA300, as well as to determine the metabolic content of its extract.
Interestingly, a previous study performed on the total extract of more than 40 plants, investigated by in vitro agar-well diffusion method against Staphylococcus aureus, Bacillus cereus, Escherichia coli, Listeria monocytogenes and Salmonella anatum, showed that plants rich in phenolics possess high antioxidant activity and hence exhibited promising antibacterial activity.23 Intriguingly, the authors showed that Gram-positive bacteria were more sensitive to the tested extracts than Gram-negative bacteria.23 Also, S. aureus was the most sensitive showing positive correlation between the antibacterial activity, antioxidant activity and the phenolic content (R2= 0.93), suggesting that the antibacterial activity was closely associated with the phenolic content.
The contribution of phenolic compounds in the antibacterial activity of Gram-positive than Gram-negative bacteria is supported by the structural differences in the outer layers. Gram-positive bacteria lack the outer membrane and the unique periplasmic space found in Gram-negative bacteria.24 The lipopolysaccharides-rich hydrophilic surface of outer membrane presents a barrier for the entry of molecules, while, the hydrolytic enzymes in the periplasmic space contribute to the breakdown of the introduced molecules. Although the mechanism of antibacterial potentials of polyphenols is not fully elucidated, there are many arguments that phenolic compounds can either i) affect the cell membrane permeability, ii) induce hydrogen-binding to intracellular enzymes and thus influence their proper function, iii) cause irreversible damages in the cytoplasmic membrane and coagulation of the cell contents and/or iv) induce integrity losses in the rigidity of cell membranes.25
Interestingly, a previous study has shown that the antioxidant activity of star anise ethanol (80%) fraction is higher than the non-polar petroleum ether fraction, owing to the solubility of phenolic compounds as well as sugars, which are known to have antioxygenic activity, in ethanol.26
The aqueous methanolic extract of star anise was tested here for its anti-biofilm activity and biofilm detachment activity at very low concentrations below the determined MBC. The concentrations tested against A. baumannii AB5057 were 1/10, 1/20 and 1/40 MBC. While the concentrations tested against MRSA USA300 were 1/40, 1/80 and 1/160 MBC. The results demonstrated that the tested extract exhibited significant inhibition of biofilm formation by both multi-drug resistant strains A. baumannii AB5057 and MRSA USA300 at all the tested concentrations. Hence, inhibition of biofilm formation without inhibiting the cell growth which in turn could reduce the risk of resistance emergence, adopting the approach of inhibiting the virulence traits and eradicating pathogenicity rather than killing the pathogen.27
Star anise extract showed remarkable detachment activity against the biofilm formed by A. baumannii AB50507 and MRSA USA300 at all the tested concentrations with significantly higher activity against MRSA USA300 than against A. baumannii AB5057. The possible mechanism underlying this effect may be due to the action of the extract on the lipophilic cellular membrane. Bacterial adherence to substrates is an initial step prior to biofilm formation. The mechanism of bacterial attachment to surfaces is thought to involve nonspecific attachments regulated by the hydrophobicity, hydrophilicity, or the charge of the material.28 Bacterial extracellular matrix components (extracellular polymeric substances, teichoic acid, and extracellular DNA) also act as glue for mutual bacterial binding, therefore promoting aggregation and adhesion of bacterial matrix.28
In the light of the in vitro results, we further tested the star anise extract to investigate if it could maintain their antibacterial activity in vivo. We selected a well-established murine model of MRSA skin infection as skin infections are one of the hallmarks of staphylococcal infections 14. These results revealed that the extract enhanced the recovery of the induced skin infection as indicated by the significant reduction of the bacterial load when compared to the control groups. These results indicate the possibility of discovery of new metabolites for management of MRSA skin infection; however,; it is necessary to investigate the mechanisms of action of phenolic compounds found in I. verum. The biologically guided fractionation of the extract will be indispensable to understand the relationship between the chemistry of phenolics and the proposed antibacterial activity.
Shikimic acid, a cyclohexane carboxylic acid derivative, was among the identified compounds in star anise extract. Shikimic acid has been reported to exert potential antibacterial activity specifically against S. aureus, possibly by destroying the cell membrane permeability after binding to membrane proteins and lipids.29 A close relative of shikimic acid, quinic acid, possess a great antibacterial activity against S. aureus via damaging the cell membrane.30 Two quinic acid derivatives, p-coumaroylquinic acid and feruloylquinic acid were detected in I. verum extract. Phenolic compounds constitute one of the most important and ubiquitous antibacterial agents. Protocatechuic acid, 3,4-dihydroxybenzoic acid, as well as caffeic acid have been also reported in this study and previous studies as a major phenolic derivative in I. verum.31 The antibacterial activity of protocatechuic acid, its ethyl ester and caffeic acid against S. aureus clinical strains have been proved either alone or synergistically with clindamycin and to a lesser extent with vancomycin.32 Rosmarinic acid, caffeic acid ester of 3-(3,4-dihydroxyphenyl) lactic acid, has been detected in our extract and has been shown to have potential antibacterial activity, especially against Gram-positive bacteria as MRSA.33,34 Rosmarinic acid exerts also potential activity against MRSA in combination with vancomycin.34
Flavonoids and their glycosides, phenolic compounds with very diverse chemistry, possess versatile health benefits reported in various epidemiological studies.35 Flavonoids are synthesized in plants in response to microbial attacks; therefore, it is not surprising that they exert in vitro and in vivo broad-spectrum antibacterial activity. Several flavonoids including catechin, apigenin, rutin, quercetin, kaempferol, isorhamnetin and luteolin, which have been detected in our analysis, have been shown to possess potent antibacterial activity by having multiple cellular targets, rather than one specific site of action.35 Eugenol, a volatile phenolic compound of the phenylpropene class found in many plant species, exerts also antibacterial activity.36 Previous studies reported that eugenol has antibacterial and anti-biofilm activity against antibiotic-resistant bacteria including A. baumannii and S. aureus.36,37 Glycosylation of eugenol was shown to increase its antibacterial potential especially against S. aureus and E. coli.37 In agreement with that, we have also identified a few eugenol glycosides; also, we have proved their structural analysis by MS/MS. To the best of our knowledge, these compounds have not been identified before from I. verum. The metabolic processes of the body usually produce an oxidative stress status with the formation of free radicals triggering body damage if occur at risky levels.38 Star anise extract showed a promising activity in agreement of the previous studies as an anti-oxidant agent due to its high content of phenolic and flavonoidal contents.38 From this study, we could suggest this polar fraction of star anise as a powerful antibiotic and antioxidant agent to be incorporated in the biopharmaceutical industries.
Conclusion
Star anise extract clearly showed significant inhibition and detachment activity against biofilm formed by the MDR and highly virulent A. baumannii AB5057 and MRSA USA300; this proposes its potential use in anti-virulence strategies against persistent infections. This activity could be attributed to its content of phenolic acids and flavonoids. Moreover, the present study also demonstrated that star anise extract can be included as a potential additive in topical antimicrobial preparations for the treatment of skin/wound infections by MRSA. This is the first systematic study to report the antibacterial activity of phenolics from star anise polar fraction against MRSA in relation to its metabolite profile. The phenolic compounds may serve as potential candidates for future in-depth studies of synergism and compatibility with commercial antibacterial agents.
Author Contributions
All authors contributed equally throughout the manuscript. All authors contributed to data analysis, drafting and revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no conflicts of interest to declare.
References
1. Wang GW, Hu WT, Huang BK, Qin LP. Illicium verum: a review on its botany, traditional use, chemistry and pharmacology. J Ethnopharmacol. 2011;136(1):10–20. doi:10.1016/j.jep.2011.04.051
2. Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016:2475067. doi:10.1155/2016/2475067
3. Liu X, Wu X, Tang J, Zhang L, Jia X. Trends and development in the antibiotic-resistance of Acinetobacter baumannii: a Scientometric Research Study (1991-2019). Infect Drug Resist. 2020;13:3195–3208. doi:10.2147/IDR.S264391
4. Hamza DA, Abd-Elsalam RM, Nader SM, Elhariri M, Elhelw R, El-Mahallawy HS. Pathways of methicillin-resistant Staphylococcus aureus in animal model: new insights regarding public health. Infect Drug Resist. 2020;13:1593–1600. doi:10.2147/IDR.S252332
5. El-Nagdy AH, Abdel-Fattah GM, Emarah Z. Detection and control of biofilm formation by staphylococcus aureus from febrile neutropenic patient. Infect Drug Resist. 2020;13:3091–3101. doi:10.2147/IDR.S259914
6. Vaughn AR, Haas KN, Burney W, et al. Potential role of curcumin against biofilm-producing organisms on the skin: a review. Phytother Res. 2017;31(12):1807–1816. doi:10.1002/ptr.5912
7. Amin M, Navidifar T, Shooshtari FS, et al. Association between biofilm formation, structure, and the expression levels of genes related to biofilm formation and biofilm-specific resistance of acinetobacter baumannii strains isolated from Burn Infection in Ahvaz, Iran. Infect Drug Resist. 2019;12:3867–3881. doi:10.2147/IDR.S228981
8. Sharifian P, Yaslianifard S, Fallah P, Aynesazi S, Bakhtiyari M, Mohammadzadeh M. Investigating the effect of nano-curcumin on the expression of biofilm regulatory genes of Pseudomonas aeruginosa. Infect Drug Resist. 2020;13:2477–2484. doi:10.2147/IDR.S263387
9. Sukumar MR, König B. Pomegranate extract specifically inhibits Clostridium difficile growth and toxin production without disturbing the beneficial bacteria in vitro. Infect Drug Resist. 2018;11:2357–2362. doi:10.2147/IDR.S163484
10. Salem MA, Juppner J, Bajdzienko K, Giavalisco P. Protocol: a fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods. 2016;12:45. doi:10.1186/s13007-016-0146-2
11. Elhosseiny NM, Elhezawy NB, Attia AS. Comparative proteomics analyses of Acinetobacter baumannii strains ATCC 17978 and AB5075 reveal the differential role of type II secretion system secretomes in lung colonization and ciprofloxacin resistance. Microb Pathog. 2019;128:20–27. doi:10.1016/j.micpath.2018.12.039
12. Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. The Lancet. 2006;367(9512):731–739. doi:10.1016/S0140-6736(06)68231-7
13. Humphries RM, Ambler J, Mitchell SL, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018;56(4):e01934–17. doi:10.1128/JCM.01934-17
14. Glatthardt T, Campos JCM, Chamon RC, et al. Small molecules produced by commensal staphylococcus epidermidis disrupt formation of biofilms by Staphylococcus aureus. Environ Microbiol. 2019;86(5). doi:10.1128/aem.02539-19
15. Mohamed MF, Abdelkhalek A, Seleem MN. Evaluation of short synthetic antimicrobial peptides for treatment of drug-resistant and intracellular Staphylococcus aureus. Sci Rep. 2016;6(1):29707. doi:10.1038/srep29707
16. Aryal S, Baniya MK, Danekhu K, Kunwar P, Gurung R, Koirala N. Total phenolic content, flavonoid content and antioxidant potential of wild vegetables from Western Nepal. Plants. 2019;8:4. doi:10.3390/plants8040096
17. Ben Said R, Hamed AI, Mahalel UA, et al. Tentative characterization of polyphenolic compounds in the male flowers of phoenix dactylifera by liquid chromatography coupled with mass spectrometry and DFT. Int J Mol Sci. 2017;18:3. doi:10.3390/ijms18030512
18. Mouwakeh A, Kincses A, Nové M, et al. Nigella sativa essential oil and its bioactive compounds as resistance modifiers against Staphylococcus aureus. Phytother Res. 2019;33(4):1010–1018. doi:10.1002/ptr.6294
19. Govindaraj Vaithinathan A, Vanitha A. WHO global priority pathogens list on antibiotic resistance: an urgent need for action to integrate One Health data. Perspect Public Health. 2018;138(2):87–88. doi:10.1177/1757913917743881
20. Patra JK, Das G, Bose S, et al. Star anise (Illicium verum): chemical compounds, antiviral properties, and clinical relevance. Phytother Res. 2020;34(6):1248–1267. doi:10.1002/ptr.6614
21. Yang JF, Yang CH, Chang HW, et al. Chemical composition and antibacterial activities of Illicium verum against antibiotic-resistant pathogens. J Med Food. 2010;13(5):1254–1262. doi:10.1089/jmf.2010.1086
22. Yang L, Wen KS, Ruan X, Zhao YX, Wei F, Wang Q. Response of plant secondary metabolites to environmental factors. Molecules. 2018;23. doi:10.3390/molecules23040762.
23. Shan B, Cai YZ, Brooks JD, Corke H. The in vitro antibacterial activity of dietary spice and medicinal herb extracts. Int J Food Microbiol. 2007;117(1):112–119. doi:10.1016/j.ijfoodmicro.2007.03.003
24. Miller SI, Salama NR. The gram-negative bacterial periplasm: size matters. PLoS Biol. 2018;16(1):e2004935. doi:10.1371/journal.pbio.2004935
25. Bouarab-Chibane L, Forquet V, Lanteri P, et al. Antibacterial Properties of Polyphenols: characterization and QSAR (Quantitative Structure-Activity Relationship) Models. Front Microbiol. 2019;10:829. doi:10.3389/fmicb.2019.00829
26. Padmashree A, Roopa N, Semwal AD, Sharma GK, Agathian G, Bawa AS. Star-anise (Illicium verum) and black caraway (Carum nigrum) as natural antioxidants. Food Chem. 2007;104(1):59–66. doi:10.1016/j.foodchem.2006.10.074
27. Imperi F, Fiscarelli EV, Visaggio D, Leoni L, Visca P. Activity and impact on resistance development of two antivirulence fluoropyrimidine drugs in Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2019;9:49. doi:10.3389/fcimb.2019.00049
28. Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5(6):580–592. doi:10.1016/j.chom.2009.05.011
29. Bai J, Wu Y, Liu X, Zhong K, Huang Y, Gao H. Antibacterial activity of shikimic acid from pine needles of cedrus deodara against Staphylococcus aureus through damage to cell membrane. Int J Mol Sci. 2015;16(11):27145–27155. doi:10.3390/ijms161126015
30. Bai J, Wu Y, Wang X, et al. In vitro and in vivo characterization of the antibacterial activity and membrane damage mechanism of quinic acid against Staphylococcus aureus. J Food Saf. 2018;38(1):e12416. doi:10.1111/jfs.12416
31. Shan B, Cai YZ, Sun M, Corke H. Antioxidant capacity of 26 spice extracts and characterization of their phenolic constituents. J Agric Food Chem. 2005;53(20):7749–7759. doi:10.1021/jf051513y
32. Miklasińska M, Kępa M, Wojtyczka RD, et al. Antibacterial activity of protocatechuic acid ethyl ester on staphylococcus aureus clinical strains alone and in combination with antistaphylococcal drugs. Molecules (Basel, Switzerland). 2015;20(8):13536–13549. doi:10.3390/molecules200813536
33. Matejczyk M, Swislocka R, Golonko A, Lewandowski W, Hawrylik E. Cytotoxic, genotoxic and antimicrobial activity of caffeic and rosmarinic acids and their lithium, sodium and potassium salts as potential anticancer compounds. Adv Med Sci. 2018;63(1):14–21. doi:10.1016/j.advms.2017.07.003
34. Ekambaram S. M N, V V, G S, B A. Synergistic effect of rosmarinic acid with antibiotics against Staphylococcus aureus and MRSA. Chinese J Pharmacol Toxicol. 2015;29:58.
35. Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. Sci World J. 2013;2013:162750. doi:10.1155/2013/162750
36. Karumathil DP, Surendran-Nair M, Venkitanarayanan K. Efficacy of Trans-cinnamaldehyde and Eugenol in Reducing Acinetobacter baumannii Adhesion to and Invasion of Human Keratinocytes and Controlling Wound Infection In Vitro. Phytother Res. 2016;30(12):2053–2059. doi:10.1002/ptr.5713
37. Resende DB, Martins H, Souza T, et al. Synthesis and in vitro evaluation of peracetyl and deacetyl glycosides of eugenol, isoeugenol and dihydroeugenol acting against food-contaminating bacteria. Food Chem. 2017;237:1025–1029. doi:10.1016/j.foodchem.2017.06.056
38. Yu C, Wei J, Yang C, Yang Z, Yang W, Jiang S. Effects of star anise (Illicium verum Hook.f.) essential oil on laying performance and antioxidant status of laying hens. Poult Sci. 2018;97(11):3957–3966. doi:10.3382/ps/pey263
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.