Back to Journals » Infection and Drug Resistance » Volume 16
In vitro Activity of Cefepime/Avibactam Against Carbapenem Resistant Klebsiella pneumoniae and Integrative Metabolomics-Proteomics Approach for Resistance Mechanism: A Single-Center Study
Authors Wen L, Luo C, Chen X, Liu T, Li X, Wang M
Received 11 May 2023
Accepted for publication 2 August 2023
Published 11 September 2023 Volume 2023:16 Pages 6061—6077
DOI https://doi.org/10.2147/IDR.S420898
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Lingjun Wen, Can Luo, Xinyi Chen, Tianyao Liu, Xianping Li,* Min Wang*
Department of Laboratory Medicine, The Second Xiangya Hospital of Central South University, Changsha, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Min Wang; Xianping Li, Department of Laboratory Medicine, The Second Xiangya Hospital of Central South University, 139 Renmin Middle Road, Changsha, 410011, People’s Republic of China, Tel +86 13298697558, Email [email protected]; [email protected]
Purpose: We aimed to evaluate the in vitro antibacterial effects of combination of cefepime/avibactam against carbapenem-resistant Klebsiella pneumonia (CRKP) and explore the resistance mechanism of FEP/AVI.
Patients and Methods: This study explored the in vitro antibacterial activities of ceftazidime/avibactam (CAZ/AVI) and cefepime/avibactam (FEP/AVI) against 40 and 76 CRKP clinical isolates. Proteomics and metabolomics were employed to investigate the resistance mechanisms of CRKP to FEP/AVI.
Results: FEP/AVI (MIC50/MIC90 0.5/4-64/4 μg/mL, resistance rate 17.1%) showed better antibacterial activity against CRKP than CAZ/AVI (MIC50/MIC90 4/4-128/4 μg/mL, resistance rate 20%) in vitro. Bioinformatics analysis showed that the differentially expressed proteins (DEPs) were enriched in alanine, aspartate and glutamate metabolism, and ribosome. Remarkably, transcriptional and translational activity-related pathways were inhibited in FEP/AVI resistant CRKP. Overlap analysis suggested that H-NS might play an important role in resistance to FEP/AVI in CRKP. The mRNA levels of DEPs-related genes (adhE, gltB, purA, ftsI and hns) showed the same trends as DEPs in FEP/AVI susceptible and resistant strains. FEP/AVI resistant isolates demonstrated stronger biofilm formation capacity than susceptible isolates. Metabolomics results showed that disturbed metabolites were mainly lipids, and adenine was decreased in FEP/AVI resistant CRKP.
Conclusion: These results indicated that H-NS, GltB and SpoT may directly or indirectly promote biofilm formation of CRKP and led to FEP/AVI resistance, but inhibited ribosomal function. Our study provides a mechanistic insight into the acquisition of resistance to FEP/AVI in Klebsiella pneumoniae.
Keywords: carbapenem-resistantKlebsiella pneumoniae, bacterial resistance, proteomics, metabolomics, cefepime/avibactam, ribosome
Introduction
Infectious diseases caused by CRKP pose a major health concern due to limited therapy and high mortality.1,2 CRKP has been included in the priority list by the World Health Organization, urgently requiring new drugs for treatment and to prevent its dissemination.3 Carriage of multiple resistance genes, including β-lactamases, renders common antibiotics ineffective against CRKP, while old drugs polymyxin and tigecycline available for this infection are restricted due to their safety profile.4,5
β-lactams (including cephalosporins) are the first-line treatment for bacterial infections,6 targeting penicillin-binding proteins (PBP) to halt the cross-linking of peptidoglycan on the cell wall, resulting in cell wall thinning, bacterial swelling, and eventual bacterial death.6,7 Hydrolysis of cephalosporins β-lactam ring by β-lactamase leads to drug inactivation and mediates resistance.8,9 A retrospective cohort study in patients with ESBL-producing E. coli and K. pneumoniae infection shows that the 30-day mortality is still 25.42%, even if they received carbapenems.10 Thus, development of β-lactamase inhibitors and their combination with β-lactam is one strategy to address this crisis. Avibactam, a diazabicyclooctanes that binds covalently and reversibly to β-lactamases,8 has been shown to inhibit Ambler class A (CTX-M and KPC), class C (AmpC) and class D (OXA-48) β-lactamases activity in vitro, restores the antibacterial activity of β-lactams.11–13 Combination of the third-generation cephalosporins ceftazidime and avibactam, an important option for the treatment of severe CRKP infections,14 was approved by the US Food and Drug Administration (FDA) in 2015 for the treatment of adults with intra-abdominal infection, urinary tract infection and hospital-acquired bacterial pneumonia.15 Compared with best available therapy, CAZ-AVI treatment was associated with a greater clinical success in CRKP bacteremia.16 However, within a short time when CAZ/AVI was introduced into clinic, some cases of CRKP resistance were reported.17,18
The fourth-generation cephalosporin cefepime is a broad-spectrum antibiotic frequently used in clinical practice but is easily degraded by many existing β-lactamases.15 Previous studies have found that cefepime in combination with polymyxin B demonstrated bactericidal activity in 24 out of 37 isolates.19 And FEP/AVI exhibited potency against CTX-M-15 producing Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae),7,20 but the antibacterial activity and resistance mechanism of this combination has not been extensively studied. Therefore, this study compared the antibacterial effects of CAZ/AVI and FEP/AVI against CRKP, and explored the resistance mechanism of FEP/AVI through proteomics and metabolomics studies.
Materials and Methods
Bacterial Isolates
A total of 76 non-repetitive CRKP isolates were selected from 2016 to 2022 in The Second Xiangya Hospital of Central South University (Changsha, Hunan, China) Table S1. All of the strains were part of the routine hospital laboratory procedure and identified by MALDI-TOF MS (Bruker Daltonik, Bremen, Germany).
Antimicrobial Susceptibility Testing
Bacterial antibiotic susceptibility was determined by Phoenix 100 system (Becton Dickinson, Franklin Lakes, USA) Tables S2 and S3. Cefepime, ceftazidime and avibactam were purchased from Meilunbio (Dalian, China). MICs of ceftazidime alone, CAZ/AVI (4 μg/mL), cefepime alone and FEP/AVI (4 μg/mL) were tested by broth microdilution method. The experiments were performed using Mueller Hinton (MH) broth (Rishi Biotech, Qingdao, China). The breakpoints of CAZ/AVI and FEP/AVI for CRKP isolates were taken from those for CAZ (resistance MIC ≥ 16 μg/mL) or FEP (resistance MIC ≥ 16 μg/mL) alone (according to CLSI guidance).21 ATCC 700603 Klebsiella pneumoniae was used as a quality control reference strains for antimicrobial susceptibility testing.22
Protein Extraction, Digestion and Tandem Mass Tag (TMT) Labeling
Strains were passaged after growing on Columbia blood agar plates for 18–24 hours at 37°C and grown under the same conditions. Three individual colonies of each isolate were randomly picked from the agar plates to prepare for proteomics. Bacterial cells were washed three times with PBS and immediately frozen with liquid nitrogen.
Samples were sonicated three times in lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail) on ice. The supernatant was collected by centrifugation at 12,000×g for 10 min at 4°C (to remove the cell debris). The protein concentration was determined with BCA kit (Beyotime, Nanjing, China).
For digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56 °C, and alkylated with 11 mM iodoacetamide in the dark at room temperature for 15 min. Then 100 mM TEAB was added to dilute the urea concentration in the protein sample to less than 2 M. Finally, samples were firstly digested (1:50 trypsin-to-protein mass ratio) overnight and incubated for a second 4h digestion (1:100 trypsin-to-protein mass ratio).
Following digestion, peptide was desalted using Strata X C18 SPE column (Phenomenex, USA) and vacuum-dried. Peptide was reconstituted in 0.5 M TEAB and processed according to the manufacturer’s protocol for TMT kit (Thermo Fisher Scientific, Waltham, USA).
HPLC Fractionation and LC-MS/MS Analysis
The tryptic peptides were separated into fractions using high pH reverse-phase HPLC with Agilent 300Extend C18 column (5 μm particles, 4.6 mm ID, 250 mm length). Briefly, peptides were first separated with a gradient of 8% to 32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Then, the peptide was combined into 18 fractions and dried by vacuum centrifuging.
After dissolved in solvent A (0.1% formic acid), the peptides were loaded onto a home-made reversed-phase analytical column (15-cm length, 75 μm i.d.). At a constant flow rate of 400 nL/min, solvent B (0.1% formic acid in 98% acetonitrile) increased from 6% to 23% over 26 min, from 23% to 35% in 8 min, increased to 80% in 3 min, and then maintained at 80% for the last 3 min on an EASY-nLC 1000 UPLC system (Thermo).
The peptides were analyzed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) with the UPLC after subjected to nanospray ion (NSI) source. Orbitrap with a resolution of 70,000 was used to detect intact peptides. Peptides were then selected for MS/MS using the NCE setting of 28 and fragments were detected at a 17,500 resolution in the Orbitrap. For data-independent acquisition, each MS scan was followed by 20 MS/MS scans with 15.0s dynamic exclusion time. Automatic gain control (AGC) was set at 5E4. The fixed first mass was set as 100 m/z.
LC-MS/MS Metabolomic Analysis
Bacterial culture was adjusted to an optical density at 600 nm (OD600) of 0.50±0.02 with normal saline to ensure the amount of cell is 1×108 CFU/mL. 100 μL bacteria solution was diluted 10 times to obtain 1×107 cells. Six biological replicates of each strain were prepared. One-milliliter diluted culture was centrifuged at 1000×g and 4 °C for 10 min to collect the cells. After removing the supernatant, bacteria were rapidly quenched in liquid nitrogen for 30s to stop the metabolic process. Bacteria were thawed on ice, washed twice with 4°C pre-cooled PBS, then centrifuged at 1000×g for 4 min at 4 °C to remove PBS. Samples were stored at −80 °C before testing.
The cellular metabolites were extracted by addition of 600 μL extract solution (acetonitrile: methanol: water = 2:2:1, with isotopically labelled internal standard mixture) three times and a subsequent 30s vortex. Whereafter, samples were ground at 35 Hz for 4 min and sonicated for 5 min (ice water bath). The homogenization and sonication were repeated 3 times. After standing at −40°C for 1h, the samples were centrifuged at 4 °C, 12,000 rpm for 15 min. The supernatant obtained was used for analysis. An equal volume of supernatant from each sample was mixed as the quality control (QC) sample.
Metabolomics analysis was performed using an ultra high-performance liquid chromatography (UHPLC) system (ThermoFisher Scientific, Waltham, USA) with a UPLC BEH Amide column (2.1 mm×100 mm, 1.7 μm, Waters, Milford Massachusetts, USA) coupled to Q Exactive HFX mass spectrometer (ThermoFisher Scientific, Waltham, USA) in both positive and negative ion modes.
The metabolites were separated by mobile phase A (25 mmol/L ammonium acetate and 25 mmol/L ammonia hydroxide in water, pH = 9.75) and mobile phase B (acetonitrile) at a flow rate of 0.5 mL/min at 25°C. Gradient elution condition was as follows: 0~0.5 min, 95%B; 0.5~7.0 min, 95%~65% B; 7.0~8.0 min, 65%~40% B; 8.0~9.0 min, 40% B; 9.0~9.1 min, 40%~95% B; 9.1~12.0 min, 95% B. The auto-sampler temperature and column temperature were 4°C and 25°C, respectively. The injection volume was 2 μL. The MS/MS spectra were acquired through QE HFX mass spectrometer (ThermoFisher Scientific, Waltham, USA) combined with software Xcalibur (Thermo). The ESI source conditions were set as follows: sheath gas flow rate, 50 Arb, Aux gas flow rate, 10 Arb, capillary temperature as 320°C, MS/MS resolution as 7500, full MS resolution as 60,000, collision energy was 10/30/60 in NCE mode. The spraying voltage was 3.5 kV (positive) or −3.2 kV (negative), respectively.
Data Preprocessing and Bioinformatic Analysis
LC-MS/MS data processing and bioinformatics analysis were performed as described in our previous study.23 The P-value (Student’s t-test) and the variable importance in the projection (VIP) of the first principal component in orthogonal projection latent structure discrimination analysis (OPLS-DA) analysis was used as a threshold to filter differential metabolites.
Reverse Transcription Quantitative PCR (RT-qPCR)
Total RNA was extracted with RNAprep pure Bacteria Kit (Tian Gen Biotech, Beijing, China). cDNA was obtained by reverse transcription with the PrimeScript RT reagent kit (Takara Bio, Shiga, Japan). A total of 5 DEPs (hns, adhE, ftsI, putA and gltB) were verified by SYBR qPCR Mix Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s protocol and analyzed with 2−ΔΔCt method. 16S rRNA was used as internal control. The primer sequences are given in Table S4.
Biofilm Formation Assay
Biofilm production was determined by crystal violet staining method. Bacterial culture was adjusted into suspension (0.5 McFarland) with sterile saline and then diluted 1:100 (1×106 CFU/mL) in Luria-Bertani (LB) broth (Sangon Biotech, Shanghai, China). For each strain, 200 μL of bacterial suspension was transferred to four wells of 96-well polystyrene plates. Each well was gently washed three times with sterile saline after static incubation at 37 °C for 24 h. Biofilms were stained for 15 min with 200 µL of 0.1% (wt/vol) crystal violet added to the desiccated wells. Then the wells were washed with sterile normal saline three times. After decolorization with 95% ethanol (v/v) for 20min, the absorbance at 595 nm was detected by Thermo Multiskan GO (ThermoFisher Scientific, Waltham, USA). The experiment was carried out in triple. The NTUH-K2044 strain, which exhibited strong biofilm formation, served as the positive control.24
Statistical Analysis
Statistical analysis was performed using Graph Pad Prism 7.0 software. Mann–Whitney U-test was used for RT-qPCR data, and the biofilm formation ability was analyzed by unpaired, two-tailed Student’s t-test. All data was displayed as mean±SD. P < 0.05 was considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001).
Results
Antimicrobial Activity of CAZ/AVI and FEP/AVI Against CRKP Isolates
In vitro activity of FEP combined with AVI at 4 μg/mL against 76 CRKP clinical isolates was determined. The antibacterial effect of CAZ/AVI on 40 CRKP strains was further confirmed (Table 1). The MIC values of FEP against the other 36 strains were also tested (Table 2). Combining 4 μg/mL AVI greatly increased the sensitivity of CRKP to FEP, reducing the MIC value by 2 to >512-fold. For the 40 CRKP isolates in which both CAZ/AVI and FEP/AVI susceptibility were determined, all FEP/AVI resistant strains were resistant to CAZ/AVI. Overall, MIC50/MIC90 of FEP/AVI (MIC50 0.5/4 μg/mL and MIC90 64/4 μg/mL) against CRKP were lower than that of CAZ/AVI (MIC50 4/4 μg/mL and MIC90 128/4 μg/mL), with 13/76 (17.1%) and 8/40 (20%) isolates being resistant (Table 3). The cumulative inhibition curve of FEP/AVI against CRKP strains shifted to the left compared with that of CAZ/AVI (Figure 1). These results suggested that the combination of FEP/AVI is more effective than CAZ/AVI.
 |
Table 1 MICs of FEP/AVI and CAZ/AVI to 40 CRKP Isolates |
 |
Table 2 MIC of FEP Alone and Combined with 4 μg/mL AVI Against 36 CRKP Isolates |
 |
Table 3 In vitro Potency of CAZ/AVI and FEP/AVI Against CRKP Clinical Isolates |
 |
Figure 1 Cumulative inhibition curves of CAZ/AVI and FEP/AVI against CRKP strains. |
Global Proteome Difference Between FEP/AVI Resistant and Susceptible K. pneumoniae
Proteomics and metabolomics were performed on a FEP/AVI resistant CRKP (indicated as CRKP in the omics results), a FEP/AVI susceptible CRKP isolate (indicated as KPWT) and a K. pneumoniae clinical isolate (SKP) susceptible to commonly used antibiotics (β-lactams, aminoglycosides, quinolones, chloramphenicol and tetracycline). The protein with fold change (FC) >1.3 or <0.77 and P-value <0.05 (Student’s t-test) of each compared group was defined as DEP. The omics data was analyzed by three comparisons, CRKP vs KPWT (Figure 2A), CRKP vs SKP (Figure 2B) and KPWT vs SKP (Figure 2C). Detailed proteomic results are presented in Dataset S1. A total of 435 DEPs were identified in CRKP vs KPWT group, of which 215 were up-regulated and 220 were down-regulated in CRKP (Figure 2D). Among these DEPs, the highest up-regulated proteins were dipeptidase D and abortive phage infection protein, which were up-regulated by 24.457-fold and 19.495-fold in CRKP, respectively; the most significant down-regulated protein was putative glycosyltransferase, which were down-regulated 0.038-fold.
Functional Enrichment of DEPs Between CRKP and KPWT
Based on GO enrichment analysis, the DEPs were mainly concentrated in oligosaccharide transport and ribonucleoprotein complex assembly in “biological process” category (Figure 3A); in “cellular component” category, the DEPs were mainly enriched in ribonucleoprotein complex, intracellular ribonucleoprotein complex and ribosome (Figure 3B); in “molecular function” category, the DEPs were mainly related to structural constituent of ribosome (Figure 3C). The affected protein domain was principally aminoglycoside phosphotransferase (Figure 3D). These results suggested that acquisition of FEP/AVI resistance in K. pneumoniae caused significant changes in ribosomal composition, structure and function.
Furthermore, KEGG enrichment analysis was conducted to reveal the pivotal regulatory pathways associated with FEP/AVI resistance. Among the significantly enriched pathways, ribosomes (S3, S4, S7, S9, S11, S14, S16, S19, S20, S21, L2, L19, L23, L24, L28, L30, L31, L32 and L34) and RNA degradation were largely inhibited in CRKP, while alanine, aspartate and glutamate metabolism (including adenylosuccinate synthetase PurA, glutamate dehydrogenase, glutamate synthase GltB, aspartate carbamoyltransferase PyrB, bifunctional protein PutA etc.) and glycerolipid metabolism were partially activated (Figure 3E). Three of the 8 enriched pathways contribute to lipid metabolism and carbon metabolism, including galactose metabolism and fatty acid degradation (including aldehyde-alcohol dehydrogenase AdhE etc.). Moreover, PBP3 (encoded by ftsI gene, FC = 0.72) and (p)ppGpp synthetase SpoT (FC = 0.768) abundance was decreased, and porin LamB (FC = 1.76) was up-regulated in CRKP. Remarkably, all 19 ribosomal proteins had decreased abundance in CRKP (Figure 4), indicating that ribosome function and transcriptional activity were remodeled in K. pneumoniae to cope with FEP/AVI stress.
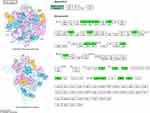 |
Figure 4 Kpn03010 ribosome pathway. Green boxed proteins indicate down-regulated proteins. |
All the DEPs were analyzed based on the Retrieval of Interaction Gene/Proteins (STRING) database for protein–protein interaction (PPI) network analysis (Figure 3F).
Co-Expressed DEPs Associated with FEP/AVI Resistance Were Screened and the Expression of Key Genes Was Verified
Due to the different patient sources of CRKP and KPWT strains used in proteomics studies, DEPs were further screened to obtain the proteins closely related to FEP/AVI resistance. We kept the co-expressed DEPs in CRKP vs KPWT group and CRKP vs SKP group, deleting the DEPs included in the KPWT vs SKP group (Figure 5A). A total of 131 DEPs were retained through the above overlap analysis (Dataset S1), among which 88 were upregulated and 43 were downregulated in CRKP (compared with KPWT). All the co-expressed DEPs had the same expression trend in both CRKP vs KPWT and CRKP vs SKP comparisons. These DEPs were mainly enriched in fatty acid degradation (including AdhE etc.), tyrosine metabolism, and alanine, aspartate and glutamate metabolism (GltB, PyrB and so on) (Figure 5B). Further narrowing the DEPs by FC > 10 or < 0.1, a total of 15 up-regulated and 1 down-regulated proteins were obtained (Table 4). AdhE and the histone-like nucleoid-structuring protein H-NS displayed the highest FC in CRKP. Transcription levels of hns, adhE, ftsI, putA and gltB (Figure 5C–G) in FEP/AVI resistant CRKP isolates (CRKP group) were consistent with their proteomic changes, compared with FEP/AVI susceptible CRKP isolates (KPWT group). Since it was previously reported that hns participated in regulating biofilm formation, biofilm formation ability was also examined. Results showed that biofilm formation ability of strains in CRKP group was significantly stronger than that of the KPWT group (P<0.0001) (Figure 5H).
 |
Table 4 Common Proteins Retained by Overlap Analysis |
Lipid Changes Occurred in FEP/AVI Resistant K. pneumoniae
To elucidate the metabolites relevant to FEP/AVI resistance in K. pneumoniae, metabolomics analysis was conducted on the three strains involved in proteomics study. The principal component analysis (PCA) score plot indicated that biological replicates of each sample were almost within the confidence interval, and differences of metabolites among CRKP, KPWT and SKP could be distinguished (Figure 6A–C). Furthermore, OPLS-DA model was established to obtain more reliable information on the difference of metabolites between groups. The permutation test for OPLS-DA model showed that the model has good predictability and predictability for the variable (Figure 6D–F). In addition, the VIP of the OPLS-DA was considered as a criterion of differentially expressed metabolites. In this study, only metabolites with VIP > 1 and P < 0.05 (Student’s t-test) were defined as differential metabolites.
Results of three comparison groups are shown in Figure 7G–I and Dataset S2. Since overlap analysis only retained a few differential metabolites, this study only explores the differential metabolites found in CRKP vs KPWT comparison. A total of 26 differential metabolites were identified, of which 10 were up-regulated and 16 were down-regulated in the CRKP (Table S5). Compared with the FEP/AVI susceptible KPWT strain, a variety of lipids were observed to be altered in CRKP, and these lipids were primarily fatty acids (pelargonic acid, heptanoic acid, stearidonic acid and linolenic acid). Most of the differential lipids were decreased in CRKP, suggesting that lipid alteration may promote the development of FEP/AVI resistance in K. pneumoniae. The differential metabolites with the largest regulation ratio (CRKP/KPWT) were C16 sphinganine (FC = 5.35) and γ-linolenic acid (FC = 0.09).
Purine Metabolism Was Affected in FEP/AVI Resistant K. pneumoniae
Pathway enrichment analysis of differential metabolites in the CRKP vs KPWT comparison was conducted. Only serine and threonine metabolism and purine metabolism were enriched (Figure 7A). Both adenine (key metabolite of purine metabolism) and betaine (differential metabolite of glycine, serine and threonine metabolism) showed reduced abundance in CRKP (Figure 7B). Comprehensive proteomic and metabolomic changes indicated that transcription and translation functions, and lipid metabolism were altered in FEP/AVI resistant K. pneumoniae (Figure 8).
Discussion
CRKP infections are challenging to treat due to limited treatment options. The combination of ceftazidime and avibactam is an important option for the treatment of CRKP infections. However, rapid rise in resistance to CAZ/AVI in K. pneumoniae calls for new β-lactam/β-lactamase inhibitor combinations for treatment. This study shows that the combination of AVI can restore the sensitivity to FEP in most CRKP strains. The in vitro antibacterial effect of this combination on CRKP is slightly better than that of CAZ/AVI, but its 17.1% resistance rate cannot be underestimated. In addition, strain 1705 was resistant to both CAZ/AVI and polymyxin (data not shown), which was susceptible to FEP/AVI. These results suggest that FEP/AVI may be a potential treatment for CRKP infection. A study published in 2012 reported that K. pneumoniae isolates carrying OXA-48 or CTX-M-15 were 100% susceptible to FEP/AVI.20 Such a marked increase in the resistance rate of K. pneumoniae to FEP/AVI during this decade suggests that the resistance of K. pneumoniae to antimicrobials has risen to an alarming level. For a deeper understanding of the mechanisms by which CRKP generates FEP/AVI resistance and to improve the effectiveness of this combination, we conducted proteomics and metabolomics studies on K. pneumoniae with different antibiotic susceptibility phenotype.
H-NS was screened out by overlap analysis, which is the global regulator of environmental and stress-responsive genes (including virulence and quorum-sensing genes).25–28 H-NS is highly conserved in Gram-negative bacteria and is a transcriptional silencer, acting by binding to AT-rich DNA regions.25,26 Several studies have shown a link between reduced antibiotic sensitivity and enhanced biofilm formation in bacteria.29,30 Studies have demonstrated the association between H-NS and biofilm formation,31,32 although the regulation of H-NS on biofilm shows opposite effects among different bacteria species.28,31,32 Bacterial biofilms are composed of its self-produced extracellular polymeric substances (EPSs), mainly including extracellular DNA (eDNA), exopolysaccharides, proteins and lipids.33 H-NS functions as an inhibitory molecule of exopolysaccharide synthesis genes in Actinobacillus pleuropneumoniae34 and Vibrio cholerae,35 whereas inactivation of hns reduced biofilm formation in E. coli,36 Acinetobacter baumannii (A. baumannii)27 and Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans).37 But the specific mechanism by which H-NS upregulation promotes biofilm formation remains unclear. The highly expressed Aldehyde-alcohol dehydrogenase ADH in FEP/AVI resistant strain was confirmed to be involved in stress resistance and regulate quorum sensing pathway during biofilm formation in A. baumannii.38 AdhE is associated with NAD+ regeneration, which is crucial for glycolysis.39 Loss of GltB impairs the ability to assimilate glutamate and reduces γ-polyglutamate biosynthesis, resulting in biofilm formation defects.40 In this study, the biofilm formation ability of FEP/AVI resistant CRKP strains was significantly stronger than that of susceptible strains, suggesting that enhanced biofilm may help CRKP resist FEP/AVI. This effect may be related to the increase of H-NS, AdhE and GltB, although the transcript level of these genes was not statistically different between FEP/AVI susceptible and resistant groups. Furthermore, the role of H-NS in bacterial resistance to antibiotics remains controversial, and it exhibits contrary regulatory functions for different bacteria and different antibiotics. H-NS can modulate bacterial multidrug resistance through the expression of efflux genes. Deletion of hns confers resistance to cloxacillin, nafcillin and cefamandole in Salmonella enterica serovar via efflux pump TolC and AcrEF.41 In contrast, although H-NS was increased in the FEP/AVI resistant CRKP in this study, expression of TolC was also upregulated, suggesting that antibiotic resistance is regulated by a complex network. Erythromycin could activate the msr(E)-mph(E) operon of K. pneumoniae and E. coli via H-NS, which confers high macrolide resistance.42 Research on carbapenem resistant A. baumannii have found that H-NS alleviated the fitness cost caused by expression of NDM-1, VIM-2 and SPM-1, and overcame the envelope stress caused by metallo-β-lactamases.26
Previous studies have found that loss of hns in A. actinomycetemcomitans can upregulate the expression of a series of proteins enriched in “regulation of translation” and “structural constituent of ribosome” items.37 Similarly, upregulation of H-NS and decreased abundance of 19 ribosomal subunits occurred in FEP/AVI resistant CRKP in this study. Given that ribosome is not the target of FEP, we speculated that the downregulation of ribosomal subunits may be associated with the upregulation of H-NS and may thereby causing fitness cost. Both H-NS and ribosomal proteins (RpsJ, RpsE, and RpsP) interacted with colistin resistance protein MCR-1 in E. coli, possibly regulating protein biosynthetic process.43 Ribosome production and correct assembly is a tightly regulated process and is essential for cell growth.44 These alterations indicated that transcriptional and translational activities in FEP/AVI resistant CRKP were inhibited. Assembly of 30S ribosome requires the exact addition of 20 proteins to the 16S ribosomal RNA.45 Ribosomal proteins S4 and S7 are initiator proteins for ribosomal assembly.46 S7 is essential for stable binding of other proteins to the 16S 3’ domain.44 S4 is one of the earliest rRNA-binding proteins involved in the assembly and folding of the 30S ribosome, which is critical for translation fidelity.45 L24 is essential for assembly of the ribosomal 50S subunit.47 Redistribution of energy by decreasing ribosomal activity may be a strategy in response to energy limitation.48 Downregulation of ribosomal metabolism and upregulation of the tricarboxylic acid cycle help Staphylococcus aureus maintain energy for survival under ciprofloxacin stress.49 Furthermore, the impaired translational function is a marker of physiological dormancy and a condition for persistent cell formation, contributing to antibiotic tolerance.50 Since it is uncertain whether the inhibition of ribosomal function is caused by H-NS, we speculate that H-NS enhances the resistance of K. pneumoniae to FEP/AVI by promoting biofilm formation and other unknown pathways, but also damages to ribosomal function. Or K. pneumoniae better adapt to antibiotic stress by downregulating translation activity.
Guanosine pentaphosphate (pppGpp) and guanosine tetraphosphate (ppGpp) are collectively referred to (p)ppGpp.51 (P)ppGpp is a major regulator of bacterial virulence, persistence, and antimicrobial resistance.51 Its synthesis is regulated by two enzymes RelA and SpoT in Gram negative bacteria.51 RelA has only synthase activity, while SpoT is a bifunctional enzyme with both synthase and hydrolase domains.52–54 But SpoT mainly exhibits weak synthetic activity and dominant (p)ppGpp hydrolase activity.52–54 Our proteomics data showed that SpoT had a decreased abundance in FEP/AVI resistant CRKP compared with susceptible strain (shown in Dataset S1), which may lead to accumulation of (p)ppGpp in bacterial cells. (P)ppGpp concentration is known to positively correlate with tolerance and resistance to β-lactams55,56 (P)ppGpp enhances β-lactam resistance in E. coli.54,57,58 (P)pGpp can also induce the expression of efflux pump genes in A. baumannii and affect antibiotic susceptibility.59 Recent studies have found that (p)ppGpp can bind to ribosome-associated GTPases (LepA, HflX, Era, and RsgA) in E. coli, therefore affecting ribosome biogenesis and translation.60 Moreover, (p)ppGpp can prevent DNA replication and elongation and greatly reduce bacterial proliferation.61 The PurA, which is involved in purine salvage pathway, was downregulated in FEP/AVI resistant strain and has been reported to be inhibited by (p)ppGpp.60,62 Decreased adenine in FEP/AVI resistant strain further implicated impaired translation function. PBP3 is one of the targets of FEP.63 It was found that PBP3 decreased in FEP/AVI resistant CRKP, suggesting that the occurrence of antimicrobial resistance may be achieved by reducing binding with antibiotics. However, there was no significant difference in the transcription level of ftsI between the FEP/AVI susceptible and resistant groups, which may be due to the insufficient number of strains used for validation. Furthermore, changes in the protein structure of PBP3 can also decrease the affinity of CRKP with FEP. But proteomics only detects the expression level and cannot obtain structural information.
Maltoporin LamB is located in the outer membrane of E. coli and K. pneumoniae, involving in the transport of maltose and maltodextrin.63,64 It was observed that deletion of lamB in K. pneumoniae resulted in modest reductions in the MICs of ceftazidime and cefepime, although resistance to carbapenems increased.65 Complement of wild-type lamB reversed the CAZ/AVI resistance acquired by induction in K. pneumoniae.63 This study found that the expression of LamB was increased in FEP/AVI resistant CRKP, suggesting that it may also be involved in the development of FEP/AVI resistance.
Disturbance of bacterial permeability barrier is thought to be a key step leading to drug resistance.66 Bacteria can modulate membrane fluidity by regulating the types and compositions of phospholipids and fatty acids, thereby maintaining the stability and normal physiological functions of cell membrane and adapting to osmotic pressure.67 Enriched fatty acid degradation pathway retained by overlap analysis and reduced fatty acid in FEP/AVI resistant strain point to cell membrane remodeling, which may be a way to resist FEP/AVI-induced osmotic stress in bacterial cells.
In conclusion, FEP/AVI resistance in CRKP is mediated by complex factors. This study highlighted the role of biofilm formation enhanced by multiple molecules and concomitant ribosome function inhibition in FEP/AVI resistance in CRKP. And future studies are needed to validate these findings and further investigate the resistance mechanisms of other antibiotics against CRKP.
This study is not without limitations. The limited number of clinical isolates limits the generalization of our results to the larger CRKP population. We conducted this study in vitro and did not investigate the clinical efficacy of FEP/AVI against CRKP infections, suggesting that more efforts need to verify if these in vitro observations may be translated to clinical utility.
Data Sharing Statement
The proteomics data is provided in Data Set 1. And the metabolomics data is available at Data Set 2. The raw proteomics data is deposited in the ProteomeXchange Consortium via the PRIDE partner repository, accession number PXD030940.
Acknowledgments
This work was supported by the Hunan Science and Technology Innovation Project under Grant (No. 2017SK50122) and the Fundamental Research Funds for the Central Universities of Central South University (2022ZZTS0949).
Disclosure
No potential conflict of interest was reported by the authors.
References
1. Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29(12):1099–1106. doi:10.1086/592412
2. Huang W, Qiao F, Zhang Y, et al. In-hospital medical costs of infections caused by carbapenem-resistant Klebsiella pneumoniae. Clin Infect Dis. 2018;67(suppl_2):S225–S230. doi:10.1093/cid/ciy642
3. World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva, Switzerland: World Health Organization; 2017.
4. Yahav D, Giske CG, Grāmatniece A, Abodakpi H, Tam VH, Leibovici L. New β-Lactam-β-Lactamase Inhibitor Combinations. Clin Microbiol Rev. 2020;34(1). doi:10.1128/CMR.00115-20
5. Katip W, Rayanakorn A, Oberdorfer P, Taruangsri P, Nampuan T. Short versus long course of colistin treatment for carbapenem-resistant A. baumannii in critically ill patients: a propensity score matching study. J Infect Public Health. 2023;16(8):1249–1255. doi:10.1016/j.jiph.2023.05.024
6. Roach EJ, Uehara T, Daigle DM, Six DA, Khursigara CM, Lainhart W. The next-generation β-lactamase inhibitor taniborbactam restores the morphological effects of cefepime in KPC-producing Escherichia coli. Microbiol Spectr. 2021;9(2):e0091821. doi:10.1128/Spectrum.00918-21
7. Everaert A, Coenye T. Effect of β-Lactamase inhibitors on in vitro activity of β-Lactam antibiotics against Burkholderia cepacia complex species. Antimicrob Resist Infect Control. 2016;5(1):44. doi:10.1186/s13756-016-0142-3
8. Ehmann DE, Jahić H, Ross PL, et al. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A. 2012;109(29):11663–11668. doi:10.1073/pnas.1205073109
9. van Haren MJ, Tehrani K, Kotsogianni I, et al. Cephalosporin prodrug inhibitors overcome metallo-β-lactamase driven antibiotic resistance. Chemistry. 2021;27(11):3806–3811. doi:10.1002/chem.202004694
10. Katip W, Yoodee J, Uitrakul S, Oberdorfer P. Efficacy of loading dose colistin versus carbapenems for treatment of extended spectrum beta lactamase producing Enterobacteriaceae. Sci Rep. 2021;11(1):18. doi:10.1038/s41598-020-78098-4
11. Ehmann DE, Jahic H, Ross PL, et al. Kinetics of avibactam inhibition against Class A, C, and D β-lactamases. J Biol Chem. 2013;288(39):27960–27971. doi:10.1074/jbc.M113.485979
12. Li H, Estabrook M, Jacoby GA, Nichols WW, Testa RT, Bush K. In vitro susceptibility of characterized β-lactamase-producing strains tested with avibactam combinations. Antimicrob Agents Chemother. 2015;59(3):1789–1793. doi:10.1128/aac.04191-14
13. Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):124.e1–124.e4. doi:10.1016/j.cmi.2019.08.020
14. Tumbarello M, Raffaelli F, Giannella M, et al. Ceftazidime-avibactam use for Klebsiella pneumoniae carbapenemase-producing K. pneumoniae infections: a retrospective observational multicenter study. Clin Infect Dis. 2021;73(9):1664–1676. doi:10.1093/cid/ciab176
15. Kloezen W, Melchers RJ, Georgiou PC, Mouton JW, Meletiadis J. Activity of cefepime in combination with the Novel β-Lactamase Inhibitor Taniborbactam (VNRX-5133) against extended-spectrum-β-lactamase-producing isolates in in vitro checkerboard assays. Antimicrob Agents Chemother. 2021;65(4). doi:10.1128/AAC.02338-20
16. Lima O, Sousa A, Longueira-Suárez R, et al. Ceftazidime-avibactam treatment in bacteremia caused by OXA-48 carbapenemase-producing Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2022;41(9):1173–1182. doi:10.1007/s10096-022-04482-9
17. Göttig S, Frank D, Mungo E, et al. Emergence of ceftazidime/avibactam resistance in KPC-3-producing Klebsiella pneumoniae in vivo. J Antimicrob Chemother. 2019;74(11):3211–3216. doi:10.1093/jac/dkz330
18. Han X, Shi Q, Mao Y, et al. Emergence of ceftazidime/avibactam and tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae due to in-host microevolution. Front Cell Infect Microbiol. 2021;11:757470. doi:10.3389/fcimb.2021.757470
19. Teo JQ, Fauzi N, Ho JJ, et al. In vitro bactericidal activities of combination antibiotic therapies against carbapenem-resistant Klebsiella pneumoniae with different carbapenemases and sequence types. Front Microbiol. 2021;12:779988. doi:10.3389/fmicb.2021.779988
20. Aktaş Z, Kayacan C, Oncul O. In vitro activity of avibactam (NXL104) in combination with β-lactams against Gram-negative bacteria, including OXA-48 β-lactamase-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2012;39(1):86–89. doi:10.1016/j.ijantimicag.2011.09.012
21. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.M100-Ed30. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
22. Khalid A, Lubián AF, Ma L, Lin RCY, Iredell JR. Characterizing the role of porin mutations in susceptibility of beta lactamase producing Klebsiella pneumoniae isolates to ceftaroline and ceftaroline-avibactam. Int J Infect Dis. 2020;93:252–257. doi:10.1016/j.ijid.2020.02.005
23. Chen X, Tian J, Luo C, Wang X, Li X, Wang M. Cell Membrane Remodeling Mediates Polymyxin B Resistance in Klebsiella pneumoniae: an Integrated Proteomics and Metabolomics Study. Front Microbiol. 2022;13:810403. doi:10.3389/fmicb.2022.810403
24. Fan LP, Yu Y, Huang S, et al. Genetic characterization and passage instability of a novel hybrid virulence plasmid in a ST23 hypervirulent Klebsiella pneumoniae. Front Cell Infect Microbiol. 2022;12:870779. doi:10.3389/fcimb.2022.870779
25. Hong SH, Wang X, Wood TK. Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli. Microb Biotechnol. 2010;3(3):344–356. doi:10.1111/j.1751-7915.2010.00164.x
26. Huang F, Fitchett N, Razo-Gutierrez C, et al. The H-NS regulator plays a role in the stress induced by carbapenemase expression in Acinetobacter baumannii. mSphere. 2020;5(4). doi:10.1128/mSphere.00793-20
27. Rodgers D, Le C, Pimentel C, et al. Histone-like nucleoid-structuring protein (H-NS) regulatory role in antibiotic resistance in Acinetobacter baumannii. Sci Rep. 2021;11(1):18414. doi:10.1038/s41598-021-98101-w
28. Behringer MG, Choi BI, Miller SF, et al. Escherichia coli cultures maintain stable subpopulation structure during long-term evolution. Proc Natl Acad Sci U S A. 2018;115(20):E4642–e4650. doi:10.1073/pnas.1708371115
29. Tang M, Wei X, Wan X, Ding Z, Ding Y, Liu J. The role and relationship with efflux pump of biofilm formation in Klebsiella pneumoniae. Microb Pathog. 2020;147:104244. doi:10.1016/j.micpath.2020.104244
30. Varadarajan AR, Allan RN, Valentin JDP, et al. An integrated model system to gain mechanistic insights into biofilm-associated antimicrobial resistance in Pseudomonas aeruginosa MPAO1. NPJ Biofilms Microbiomes. 2020;6(1):46. doi:10.1038/s41522-020-00154-8
31. Ares MA, Fernández-Vázquez JL, Rosales-Reyes R, et al. H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front Cell Infect Microbiol. 2016;6:13. doi:10.3389/fcimb.2016.00013
32. Yu Z, Zhang J, Ding M, et al. SspA positively controls exopolysaccharides production and biofilm formation by up-regulating the algU expression in Pseudoalteromonas sp. R3. Biochem Biophys Res Commun. 2020;533(4):988–994. doi:10.1016/j.bbrc.2020.09.118
33. Kim S, Li XH, Hwang HJ, Lee JH, Ercolini D. Thermoregulation of pseudomonas aeruginosa biofilm formation. Appl Environ Microbiol. 2020;86(22). doi:10.1128/AEM.01584-20
34. Bossé JT, Sinha S, Li MS, et al. Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. J Bacteriol. 2010;192(9):2414–2423. doi:10.1128/jb.01513-09
35. Wang H, Ayala JC, Silva AJ, Benitez JA. The histone-like nucleoid structuring protein (H-NS) is a repressor of Vibrio cholerae exopolysaccharide biosynthesis (vps) genes. Appl Environ Microbiol. 2012;78(7):2482–2488. doi:10.1128/aem.07629-11
36. Belik AS, Tarasova NN, Khmel IA. Регулирование образования биопленок в Escherichia coli K12: влияние мутаций в генах овв, StpA, lon и rpoN [Regulation of biofilm formation in Escherichia coli K12: effect of mutations in HNS, StpA, lon, and rpoN genes]. Mol Gen Mikrobiol Virusol. 2008;4:3–5. Russian.
37. Bao K, Bostanci N, Thurnheer T, et al. Aggregatibacter actinomycetemcomitans H-NS promotes biofilm formation and alters protein dynamics of other species within a polymicrobial oral biofilm. NPJ Biofilms Microbiomes. 2018;4(1):12. doi:10.1038/s41522-018-0055-4
38. Lin GH, Hsieh MC, Shu HY. Role of iron-containing alcohol dehydrogenases in Acinetobacter baumannii ATCC 19606 stress resistance and virulence. Int J Mol Sci. 2021;22(18):9921.
39. Pony P, Rapisarda C, Terradot L, Marza E, Fronzes R. Filamentation of the bacterial bi-functional alcohol/aldehyde dehydrogenase AdhE is essential for substrate channeling and enzymatic regulation. Nat Commun. 2020;11(1):1426. doi:10.1038/s41467-020-15214-y
40. Zhou H, Luo C, Fang X, et al. Loss of GltB inhibits biofilm formation and biocontrol efficiency of bacillus subtilis Bs916 by altering the production of γ-polyglutamate and three lipopeptides. PLoS One. 2016;11(5):e0156247. doi:10.1371/journal.pone.0156247
41. Nishino K, Hayashi-Nishino M, Yamaguchi A. H-NS modulates multidrug resistance of Salmonella enterica serovar Typhimurium by repressing multidrug efflux genes acrEF. Antimicrob Agents Chemother. 2009;53(8):3541–3543. doi:10.1128/aac.00371-09
42. Duan Y, Liu S, Gao Y, Zhang P, Mao D, Luo Y. Macrolides mediate transcriptional activation of the msr(E)-mph(E) operon through histone-like nucleoid-structuring protein (HNS) and cAMP receptor protein (CRP). J Antimicrob Chemother. 2022;77(2):391–399. doi:10.1093/jac/dkab395
43. Li H, Wang Y, Chen Q, et al. Identification of functional interactome of colistin resistance protein MCR-1 in Escherichia coli. Front Microbiol. 2020;11:583185. doi:10.3389/fmicb.2020.583185
44. Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455(7217):1268–1272. doi:10.1038/nature07298
45. Kim H, Abeysirigunawarden SC, Chen K, et al. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506(7488):334–338. doi:10.1038/nature13039
46. Nowotny V, Nierhaus KH. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27(18):7051–7055. doi:10.1021/bi00418a057
47. Nishi K, Morel-Deville F, Hershey JW, Leighton T, Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988;336(6198):496–498. doi:10.1038/336496a0
48. Müller AL, Gu W, Patsalo V, Deutzmann JS, Williamson JR, Spormann AM. An alternative resource allocation strategy in the chemolithoautotrophic archaeon Methanococcus maripaludis. Proc Natl Acad Sci U S A. 2021;118(16):e2025854118.
49. Liu J, Wei Q, Wang Z, Sun X, He QY. Proteomic Study of the adaptive mechanism of ciprofloxacin-resistant Staphylococcus aureus to the host environment. J Proteome Res. 2022;21(6):1537–1547. doi:10.1021/acs.jproteome.2c00140
50. Tomlinson BR, Malof ME, Shaw LN. A global transcriptomic analysis of Staphylococcus aureus biofilm formation across diverse clonal lineages. Microb Genom. 2021;7(7):e000598.
51. Das B, Bhadra RK. (p)ppGpp metabolism and antimicrobial resistance in bacterial pathogens. Front Microbiol. 2020;11:563944. doi:10.3389/fmicb.2020.563944
52. De Boer HA, Bakker AJ, Gruber M. Breakdown of ppGpp in spoT and spoT-cells of Escherichia coli. manganese and energy requirement and tetracycline inhibition. FEBS Lett. 1977;79(1):19–24. doi:10.1016/0014-5793(77)80341-4
53. Fitzsimmons LF, Liu L, Kant S, et al. SpoT induces intracellular salmonella virulence programs in the phagosome. mBio. 2020;11(1):10–128.
54. Kim HY, Go J, Lee KM, Oh YT, Yoon SS. Guanosine tetra- and pentaphosphate increase antibiotic tolerance by reducing reactive oxygen species production in Vibrio cholerae. J Biol Chem. 2018;293(15):5679–5694. doi:10.1074/jbc.RA117.000383
55. Spira B, Ospino K. Diversity in E. coli (p)ppGpp Levels and Its Consequences. Front Microbiol. 2020;11:1759. doi:10.3389/fmicb.2020.01759
56. Wu J, Long Q, Xie J. (p)ppGpp and drug resistance. J Cell Physiol. 2010;224(2):300–304. doi:10.1002/jcp.22158
57. Ishiguro EE, Ramey WD. Inhibition of in vitro peptidoglycan biosynthesis in Escherichia coli by guanosine 5’-diphosphate 3’-diphosphate. Can J Microbiol. 1980;26(12):1514–1518. doi:10.1139/m80-253
58. Hugonnet JE, Mengin-Lecreulx D, Monton A, et al. Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. Elife. 2016;5. doi:10.7554/eLife.19469
59. Jung HW, Kim K, Islam MM, Lee JC, Shin M. Role of ppGpp-regulated efflux genes in Acinetobacter baumannii. J Antimicrob Chemother. 2020;75(5):1130–1134. doi:10.1093/jac/dkaa014
60. Zhang Y, Zborníková E, Rejman D, Gerdes K, Swanson MS. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio. 2018;9(2). doi:10.1128/mBio.02188-17
61. Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128(5):865–875. doi:10.1016/j.cell.2006.12.043
62. Stayton MM, Fromm HJ. Guanosine 5’-diphosphate-3’-diphosphate inhibition of adenylosuccinate synthetase. J Biol Chem. 1979;254(8):2579–2581. doi:10.1016/S0021-9258(17)30108-4
63. Guo Y, Liu N, Lin Z, et al. Mutations in porin LamB contribute to ceftazidime-avibactam resistance in KPC-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2021;10(1):2042–2051. doi:10.1080/22221751.2021.1984182
64. Klebba PE, Hofnung M, Charbit A. A model of maltodextrin transport through the sugar-specific porin, LamB, based on deletion analysis. EMBO j. 1994;13(19):4670–4675. doi:10.1002/j.1460-2075.1994.tb06790.x
65. García-Sureda L, Juan C, Doménech-Sánchez A, Albertí S. Role of Klebsiella pneumoniae LamB Porin in antimicrobial resistance. Antimicrob Agents Chemother. 2011;55(4):1803–1805. doi:10.1128/aac.01441-10
66. Zgurskaya HI, Rybenkov VV. Permeability barriers of Gram-negative pathogens. Ann N Y Acad Sci. 2020;1459(1):5–18. doi:10.1111/nyas.14134
67. Jia FF, Pang XH, Zhu DQ, Zhu ZT, Sun SR, Meng XC. Role of the luxS gene in bacteriocin biosynthesis by Lactobacillus plantarum KLDS1.0391: a proteomic analysis. Sci Rep. 2017;7(1):13871. doi:10.1038/s41598-017-13231-4
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






