Back to Journals » Cancer Management and Research » Volume 10
IL-17 and colorectal cancer risk in the Middle East: gene polymorphisms and expression
Authors Al Obeed OA, Vaali-Mohammed MA, Alkhayal KA, Bin Traiki TA, Zubaidi AM, Arafah M , Harris RA , Khan Z , Abdulla MH
Received 1 January 2018
Accepted for publication 9 April 2018
Published 5 September 2018 Volume 2018:10 Pages 2653—2661
DOI https://doi.org/10.2147/CMAR.S161248
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Omar A Al Obeed,1 Mansoor-Ali Vaali-Mohamed,1 Khayal A Alkhayal,1 Thamer A Bin Traiki,1 Ahmad M Zubaidi,1 Maha Arafah,2 Robert A Harris,3 Zahid Khan,4,* Maha-Hamadien Abdulla1,*
1Colorectal Research Chair, Department of Surgery, King Khalid University Hospital, College of Medicine, King Saud University, Riyadh, Saudi Arabia; 2Department of Pathology, King Khalid University Hospital, College of Medicine, King Saud University, Riyadh, Saudi Arabia; 3Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden; 4Genome Research Chair, Department of Biochemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
*These authors contributed equally to this work
Background: IL-17 expressed by Th17 cells play a crucial role in tissue inflammation by induction of proinflammatory and neutrophil mobilizing cytokines, and IL-17 polymorphisms are associated with colorectal cancer (CRC).
Objective: We investigated the expression of IL-17 and the association of IL-17 gene polymorphisms with CRC susceptibility in a Middle East population.
Materials and methods: The study included 117 diagnosed CRC patients and 100 age- and gender-matched healthy controls. IL-17A rs2275913 (G197A) and IL-17F rs763780 (T7488C) single nucleotide polymorphisms, mRNA, and protein levels of IL-17A were assessed.
Results: We observed significant association between rs2275913 in IL-17A and susceptibility to CRC (p = 0.016228). The AG and AA genotypes conferred 2-fold and 2.8-fold, respectively, higher risk of developing CRC compared with individuals having GG genotype. Stratification of the data based on gender and age revealed very strong association of CRC with IL17A rs2275913 only in males and “AG” genotype in patients ≤57 years of age at the time of disease diagnosis. The rs763780 in IL-17F was not linked with CRCs in our cohort. Furthermore, IL-17A mRNA expression in CRCs was significantly elevated compared to adjacent normal tissues, particularly in early stages of disease (p = 0.0005). Strong immunoreactivity to IL-17A protein was observed in 70% of early stage relative to 30% of late-stage tumors.
Conclusion: The IL-17A G197A variant may be utilized as a genetic screening marker in assessing CRC risk, and its expression can be used as a biomarker for early detection of CRC in the Saudi population.
Keywords: interleukin (IL), genetic polymorphism, colorectal cancer, Saudi population
Introduction
Cancer is the leading cause of death worldwide, representing a major threat to public health. According to the latest global statistics in 2015, there were ~14.1 million new cases and 8.2 million cancer-related deaths.1 Colorectal cancer (CRC) ranks third in the USA and fourth globally in cancer-related deaths.2 In Saudi Arabia, CRC is the most common cancer type in men and the second most common in women.3
Cytokines are important inflammatory mediators of cancer growth and invasion. Th17 cells were initially described as CD4+ T cells that play pivotal roles in both adaptive and innate immunity by secreting the proinflammatory cytokine IL-17.4,5 IL-17 is the major cytokine produced by Th17 cells and induces the production of inflammatory cytokines and chemokines by neutrophils and macrophages, which may play a crucial role in human malignancies.6 IL-17 has six family members (IL17A–F).7 IL-17A and F are expressed by all subsets of Th17 cells.8 Both IL-17A and IL-17F genes reside on chromosome 6p12 and share strongest homology.9
We chose the IL-17 gene as a candidate because it is well documented that IL-17 family members play an active role in inflammation, autoimmune diseases, and cancer.10 Several studies have indicated that IL-17 is associated with CRC and gastric cancer.11,12 Increased expression of IL-17A has been reported in CRC patients.13 In contrast, excessive production of IL-17F inhibits colon tumorigenesis in human and animal CRC models. This protective role of IL17F has been attributed to its inhibiting tumor angiogenesis.14 Furthermore, genetic variants in IL-17A and IL-17F are associated with the development and progression of CRCs.15 The rs2275913 (G197A) single nucleotide polymorphism (SNP) in IL-17A gene promoter and IL-17F rs763780 (T7488C) SNP at the start codon of IL-17F gene are reported to be significantly associated with gastric cancer, breast cancer, ulcerative colitis, and Crohn’s disease in different populations.16–18
To the best of our knowledge, no study has been conducted in the Middle East addressing the relationship between IL-17A rs2275913 and IL-17F rs763780 polymorphisms and CRC risk. We therefore conducted this study to explore the possible association between IL-17A and IL-17F polymorphisms and risk of CRC in the Saudi Arabian population. Additionally, given the role of IL-17A in inflammatory and cancerous conditions, we examined its mRNA and protein expression in colorectal tumor and adjacent normal tissues.
Materials and methods
Study population
A total of 217 blood samples were obtained from individuals attending the King Khalid University Hospital. These incorporated 117 patients with CRC and 100 healthy controls. All blood samples were collected before surgery. Patients who had undergone preoperative irradiation or chemotherapy were excluded from the study. All patients and controls were Saudi Arabian, gender- and age-matched, and had been recruited after physical examinations and diagnostic exclusion of cancer.
DNA extraction
Blood samples (3 mL) from all subjects enrolled in the study were collected in EDTA-containing vacutainers. Genomic DNA was thereafter extracted using the QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA, USA), quantitated spectrophotometrically using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and stored at –80°C until assay.
SNP selection and genotyping
The IL-17A (G197A) and IL-17F (T7488C) SNPs were selected from SNP500 Cancer project and previous literatures19–21 and genotyped using a TaqMan allelic discrimination assay.22 For each sample, 5 ng of DNA per reaction was used with 12.5 µL of 2X Universal Master Mix and 200 nM of primers (Thermo Fisher Scientific). All genotypes were determined by end point reading using a ViiA™ 7 real-time PCR System (Thermo Fisher Scientific). Five percent of the samples were randomly selected and subjected to repeat analysis as a measure for verification of genotyping procedures. The results were reproducible without any discrepancies.
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
RNA from CRC and adjacent normal tissue was extracted using Trizol method and reverse transcribed to cDNA using the High Capacity cDNA kit (cat no 4368814; Applied Biosystems, Foster City, CA, USA). Quantitative PCR analysis was performed on ViiA™ 7 real-time PCR system (Thermo Fisher Scientific) using the SYBR Green PCR Master Mix (cat no 4385612; Thermo Fisher Scientific). The relative RNA level of IL-17A was normalized against GAPDH for qRT-PCR to determine IL-17A expression in patients’ tumor and adjacent normal tissue. The following primers were used: human IL-17A, 5′-CTCATTGGTGTCACTGCTACTG-3′ (sense) and 5′-CCTGGATTTCGTGGGATTGTG -3′ (antisense); human GAPDH, 5′-ACCCATCACCATCTTCCAGGAG-3′ (sense) and 5′-GAAGGGGCGGAGATGATGAC-3′ (antisense). Results were normalized to GAPDH levels using the ΔCt method, where the relative expression is calculated as 2ΔCt, and Ct represents the threshold cycle.
Immunohistochemistry
Immunohistochemical staining was conducted on 5 µm sections of tissue microarray (TMA) blocks as previously described.23 IL17A staining was performed using a streptavidin-biotinylated horseradish peroxidase (S-ABC) kit (Novo Link Max Polymer Detection System; Novocastra) involving quenching of endogenous peroxidase activity with 3% hydrogen peroxide, blocking of nonspecific binding of antibodies (Novocastra) prior to incubation first with human IL-17 (H-132) polyclonal rabbit antibody (sc-7927) (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at a dilution of 1:100 and then with biotinylated anti-rabbit immunogloublin G (Novocastra). Peroxidase was detected using diaminobenzedine substrate (Novocastra), and slides were counterstained with Mayer’s hematoxylin (Novocastra). Omission of the primary antibody was employed as a negative control. The expression of IL17A in tumor and normal samples was analyzed using the eSlide capture device (ScanScope® CS).
Image analysis
High-resolution, whole-slide digital scans of all TMA glass slides were created using a ScanScope slide scanner (Aperio Technologies, Inc.) as previously described.23
Statistical analysis
Genotype and allelic frequencies in the CRCs and healthy individuals were compared by Pearson’s goodness-of-fit chi-square tests. Chi-square values, ORs, 95% CIs, and p-values were calculated using the Web-based programs (http://ihg.gsf. de/cgi-bin/hw/hwa1.pl and http://www.socscistatistics.com). A p-value of <0.05 was considered as significant.
Bonferroni’s correction for multiple comparisons was applied with an α = 0.025 considered as significant.
Statistical analysis was performed using GraphPad Prism software (version 5.0). The mean between the two groups was compared using Mann–Whitney test and paired t-test for IL-17A mRNA levels in early and late tumor stages. P-value of ≤0.05 was considered as significant.
Ethics statement
The study has been approved by the Ethics Committee at King Khalid University Hospital, King Saud University. Patients’ written informed consents were obtained for this study.
Results
CRC risk association with IL-17A and IL-17F genotypes and alleles
The genotypes and allele frequencies of IL-17A (rs2275913) and IL-17F (rs763780) assessed in the peripheral blood cell DNA from CRC patients and healthy controls are summarized in Table 1. The distribution of three genotypes of rs2275913 (GG, AG, and AA) was significantly different in the normal healthy controls (n = 100) compared to the CRC patients (n = 117) (χ2 = 8.242, df = 2, p = 0.016228). The homozygous G allele of rs2275913 was used as the reference in order to assess the risk of acquiring CRCs in individuals with AG and AA genotypes. It was determined that in CRC patients, significantly higher proportion of AG (34.2%) and AA (14.5%) genotypes were detected compared to 23% and 7%, respectively, in the control group. It was noted that the heterozygous AG genotype and homozygous AA genotype of IL-17A conferred 2-fold and 2.8-fold, respectively, higher risk of developing CRC in individuals compared to those having the GG genotype (AG genotype, OR = 2.029, χ2 = 5.11, p = 0.02380; AA genotype, OR = 2.833, χ2 = 4.94, p = 0.02630). We also observed that similar to the rs2275913 genotypes, the distribution of allelic frequencies in CRCs was significantly different than in healthy controls (OR = 2.038, χ2 = 9.76, p = 0.00179). Furthermore, we observed that the AG genotype and the A allele of IL-17A rs2275913 SNP exhibited significant association with CRC, even after applying Bonferroni’s correction for multiple testing.
The IL-17F rs763780 SNP was not significantly associated with CRC in our study population. The distribution of genotype and allele frequencies of rs763780 was exactly similar between the CRC cases and healthy controls (Table 1). In both the study groups (CRC and controls), CC and CT genotypes were observed at a frequency of 0.94 and 0.06, respectively, whereas the homozygous TT genotype was absent in both the cases and control populations.
IL-17A/F SNPs and CRC risk age at disease diagnosis and gender-based analysis
To examine the association of IL-17A/F SNPs with the age of onset of CRC, patients were classified based on the median age at disease diagnosis as ≤57 (n = 58) and >57 (n = 59) years. The distribution of genotype and allele frequencies determined using the DNA from blood in the CRC group and age-matched normal control subjects without prior history of any cancer was compared and the analysis is presented in Table 2. The AG genotype of rs2275913 in IL-17A conferred a statistically significant risk of developing CRC at an early age of ≤57 years (OR = 2.911, χ2 = 5.03, p = 0.02486). Thus, individuals that are AG heterozygous at rs2275913 SNP are at about 3-fold higher risk of developing CRC at an age ≤57 years compared to those having the GG genotype. Additional analysis based on the allelic model suggests that the A allele of rs2275913 was significantly associated with development of CRCs, although this association does not affect the age of disease onset as the risk was observed in both age groups. As evident in the overall analysis, the IL-17F rs763780 SNP was not associated with CRC in either age group.
In order to assess whether gender plays any role in the association of IL-17A rs2275913 and IL-17F rs763780 SNPs with CRC risk, cases and control subjects were gender stratified. The distributions of genotype and allele frequencies along with statistical analysis are presented in Table 3. Interestingly, the AG and AA genotypes of rs2275913 had a statistically strong association with CRC in males (AG genotype, OR = 2.938, χ2 = 8.06, p = 0.00452; AA genotype, OR = 19.710, χ2 = 13.42, p = 0.00025). Furthermore, examination of allelic distribution revealed that the A allele of rs2275913 was prevalent at a higher proportion in CRC (38%) than in controls (14%). It was noted that the A allele of rs2275913 exerts about 4-fold higher risk of developing CRCs in males (OR = 3.746, χ2 = 20.09, p = 7.394e−06). The IL-17A rs2275913 in females and the IL-17F rs763780 polymorphism in both genders were not associated with CRC in our cohort.
IL-17A mRNA increases in early stage of CRC patients
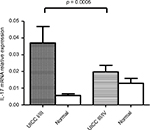
IL-17A mRNA levels were analyzed in a series of human paired specimens at early stage (I&II; n = 40) and late-stage (III&VI; n = 34) CRC. The mRNA levels in the early stage of CRC compared with the late stage were 0.037 ± 0.005 and 0.02 ± 0.01, respectively. Statistical analysis indicated that the IL-17 expression was significantly higher in CRC compared with adjacent normal tissue (p < 0.05). A significant difference was observed between early stage I/II and late stage III/IV (p = 0.0005) (Figure 1). These findings indicate that IL-17A mRNA was significantly higher in early-stage CRC.
Immunostaining of IL-17A
We performed immunohistochemical analysis of IL-17A on TMAs of paired tumor and adjacent normal specimens (n = 20). Early-stage tumor tissues contained many IL-17A-positive tumor cells compared with late-stage tumor tissues (Figure 2A and B). Moreover, epithelial cells in normal colorectal tissues were negative (Figure 2C). We performed a semi-quantitative analysis based on the staining intensity of IL-17A as negative, weak, or strong staining. According to this criterion, 70% of early-stage CRC samples showed strong positive staining, whereas only 30% samples showed strong staining in late stages. IL-17A staining was significantly different between early- and late-stage CRCs (p = 0.0001). These results demonstrate that IL-17A expression was significantly increased at the transcription and translation levels in early-stage CRC.
Discussion
There has been an increasing interest in investigating the polymorphisms of genes underlying susceptibility to tumorigenesis. Owing to its proinflammatory property, IL-17 plays an important role in the pathogenesis of many chronic inflammatory diseases, such as asthma, cystic fibrosis, chronic obstructive pulmonary disease, and inflammation-associated cancer.24 SNPs in IL-17 may alter gene expression, leading to an alteration in protein function and contributing to cancer risk.25,26 Immune coordination between T-helper subsets has been reported within the tumors, such as Th1 being cytotoxic and having favorable outcome, whereas Th17 clusters are associated with poor prognosis in CRC.27 Moreover, IL-17-producing cells may assist in the development of CRC by nurturing angiogenesis via promoting VEGF production from cancer cells. IL-17 is thus proposed as both a novel indicator and a therapeutic target in CRC patients.28 While several previous studies have reported associations between genetic polymorphisms of IL-17 and the susceptibility of CRC, the results have been inconsistent.
IL-17A, a proinflammatory cytokine involved in the etiology of several inflammatory and autoimmune diseases, is the most studied ligand of the IL-17 family.29,30 Recently, Dai et al reported in a meta-analysis that IL-17A as well as IL-17F polymorphisms increase the risk of cancer development.31 Another recent meta-analysis reported the association of IL-17A polymorphisms with an increased risk of cancer, especially breast, ovarian, and cervical cancers, in an Asian population.32 Moreover, Espinoza et al reported that peripheral blood mononuclear cells from healthy individuals carrying the 197A allele of IL-17A rs2275913 when stimulated in vitro transcribed higher levels of mRNA as well as secreted significantly more IL-17 protein compared to those not having the 197A allele. They also noted higher luciferase activity with the 197A allele than the 197G allele of IL-17A rs2275913.33 Although IL-17A and IL-17F are highly homologous and have some overlapping functions, IL-17A has ~10-fold greater strength in induction of chemokine responses.34 In view of these crucial functions of IL-17, we examined the distributions of genotypes and alleles for IL-17A and IL-17F in a cohort of CRC patients and normal healthy controls, as well as mRNA and protein expressions in colorectal cancerous and adjacent normal tissues.
Our case–control study interrogates the association between IL-17A G197A (rs2275913) and IL-17F (rs763780) polymorphism in the IL-17 gene in CRC in the Saudi population. We observed that individuals carrying the AG/AA genotypes and the A allele of IL-17A G197A SNP had an increased risk of CRC compared with those harboring the wild-type GG genotype. IL-17F (rs763780) did not exhibit a strong association. We observed a significant association of low frequency of the minor allele A of IL-17A G197A SNP in the normal control samples (0.18), compared to in CRC samples (0.31), indicating that this allele could be a risk factor in the Saudi population. This significant risk association was even observed after employing the Bonferroni correction. Interestingly, we observed a higher frequency of the A allele IL-17A G197A in male CRC samples (0.38) compared to in normal controls (0.14). This indicates that males carrying the A allele could be at higher risk of developing CRC, and this is a novel observation in our population.
Our observation concurs with a recent report that demonstrated significantly higher risk of developing CRC in individuals possessing the AG genotype of IL-17A rs2275913 in an Iranian population.35 It is also in agreement with the results that the IL-17A gene A allele is involved in susceptibility of CRC and suggested that IL-17A polymorphism may serve as a biomarker of CRC treatment outcome and progression in Tunisian patients.15,36 A previous study also reported a positive correlation between AA genotype of IL-17A G197A polymorphism and increased risk of developing gastric cancer in a Chinese population,37 although Wu et al did not find similar association with gastric cancer in the overall analysis in Chinese patients.13 Han et al described IL-17A G-197A being closely associated with susceptibility to the development of osteoarthritis in the Korean population.38 These discrepancies could be explained by differences in patient demographics, ethnicities, genetic background, distinct environmental exposures, sample selection, and sample size, all of which are known to affect association studies.39 No association between IL-17F (rs763780) polymorphism and CRC susceptibility was observed in our population. This is in agreement with a recent meta-analysis study40 that provides evidence that the IL-17A (-197G/A) polymorphism but not the IL-17F (rs763780) polymorphism might be associated with cancer risk. Furthermore, IL-17A (rs2275913) has been shown to be associated with lung and laryngeal cancer, whereas IL-17F (rs763780) was not linked with these cancers.41,42 Our results should be confirmed in more extensive studies using larger cohort, which is the core limitation of this study from different Middle Eastern population.
The rs2275913 SNP is located at –197 position from the start codon of the IL-17A gene and has the potential to influence its mRNA transcription due to the possibility of the variant being in the promoter region.43 We investigated IL-17A mRNA expression levels in CRC tissues. Increased expression was recorded in the patients’ CRC samples compared to the adjacent normal tissues. A significant difference was observed between mRNA levels in early compared with late stages of the disease, suggesting deregulation of IL-17A as an early event in colorectal carcinogenesis. This result is in agreement with previous reports in a mouse model and in human CRC samples relative to normal adjacent tissues.44,45 In contrast, Wägsäter et al did not find IL-17 protein levels to be significantly different between cancer and adjacent normal tissues in Swedish patients. They also examined IL-17 in the plasma of CRC patients and healthy controls and found no difference. Furthermore, no correlation between IL-17 expression and clinical characteristics was observed.46
Similar to mRNA, our analysis of IL-17A immunohistochemical staining of CRC tissues revealed increased expression in early-stage compared with late-stage tumors. Our results imply that IL-17A mRNA and protein expression may serve as diagnostic marker to differentiate between early and late stages of CRC and that IL-17A polymorphisms might be used as a screening marker to identify individuals with higher risk of developing CRC. IL-17A is a component of complex cytokine (IL-6 and TNF-α)47 and chemokine networks, 48 and hence involvement of other factors in the network cannot be ruled out. Due to the implication of IL-17A in the pathogenesis of several inflammation-related diseases including cancer, several strategies are under investigation to therapeutically modulate this molecule.49 Higher levels of IL-17A promotes rituximab resistance and serves as useful prognostic marker predicting unfavorable survival in patients with diffuse large B-cell lymphoma.50 Recently, Zhu et al demonstrated that andrographolide, an anti-inflammatory compound isolated from Andrographis paniculata, can decrease the levels of IL-17A among other proinflammatory factors and suggested its therapeutic potential against ulcerative colitis.51
Conclusion
Our study provides compelling evidence that the AA/AG genotype and A allele of IL-17A G197A may be associated with a higher risk of developing CRC in the Saudi population. However, no association was identified between the IL-17F rs763780 polymorphisms and the risk of CRC. Furthermore, our results provide evidence that IL-17A mRNA and protein expressions are upregulated in CRC in Saudi Arabian patients. Additional validation studies with larger sample size are required to verify the role of IL-17A as a genetic screening marker in assessing CRC risk.
Acknowledgment
The study was funded by King Saud University, through Vice Deanship of Research Chairs.
Disclosure
The authors report no conflicts of interest in this work.
References
Fitzmaurice C, Dicker D, Pain A, et al. The global burden of cancer 2013. JAMA Oncol. 2015;3(4):505–527. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Mosli MH, Al-Ahwal MS. Colorectal cancer in the Kingdom of Saudi Arabia: need for screening. Asian Pac J Cancer Prev. 2012;13(8):3809–3813. | ||
Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. | ||
Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–1183. | ||
Harrington LE, Hatton RD, Mangan PR, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6(11):1123–1132. | ||
Shi Y, Lin H, Cui J, et al. The role of interleukin-17A in colorectal tumorigenesis. Cancer Biother Radiopharm. 2013;28(6):429–432. | ||
Moseley TA, Haudenschild DR, Rose L, Reddi AH. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2003;14(2):155–174. | ||
Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282(9):5969–5972. | ||
Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21(4):467–476. | ||
Xu BL, Li YT, Dong SX, et al. IL-17 rs2275913 genetic variation contributes to the development of gastric cancer in a Chinese population. Genet Mol Res. 2016;15(2):1–7. | ||
Hou C, Yang F. Interleukin-17A gene polymorphism is associated with susceptibility to gastric cancer. Int J Clin Exp Pathol. 2015;8(6):7378–7384. eCollection 2015. | ||
Wu X, Zeng Z, Chen B, et al. Association between polymorphisms in interleukin-17A and interleukin-17F genes and risks of gastric cancer. Int J Cancer. 2010;127(1):86–92. | ||
Tong Z, Yang XO, Yan H, et al. A protective role by interleukin-17F in colon tumorigenesis. PLoS One. 2012;7(4):e34959. | ||
Omrane I, Marrakchi R, Baroudi O, et al. Significant association between interleukin-17A polymorphism and colorectal cancer. Tumour Biol. 2014;35(7):6627–6632. | ||
Kawaguchi M, Takahashi D, Hizawa N, et al. IL-17F sequence variant (His161Arg) is associated with protection against asthma and antagonizes wild-type IL-17F activity. J Allergy Clin Immunol. 2006;117(4):795–801. | ||
Büning C, Schmidt HJ, Molnar T, et al. Heterozygosity for IL23R p. Arg381Gln confers a protective effect not only against Crohn’s disease but also ulcerative colitis. Aliment Pharmacol Ther. 2007;26(7):1025–1033. | ||
Xie Y, Sheng W, Xiang J, Ye Z, Yang J. Interleukin-17F suppresses hepatocarcinoma cell growth via inhibition of tumor angiogenesis. Cancer Invest. 2010;28(6):598–607. | ||
Qi WT, Gao JL, Zhang SS. Role of IL-17 gene polymorphisms in the susceptibility to gastric cancer. Genet Mol Res. 2015;14(4):13364–13369. | ||
Wang N, Yang J, Lu J, et al. IL-17 gene polymorphism is associated with susceptibility to gastric cancer. Tumour Biol. 2014;35(10):10025–10030. | ||
Qinghai Z, Yanying W, Yunfang C, Xukui Z, Xiaoqiao Z. Effect of interleukin-17A and interleukin-17F gene polymorphisms on the risk of gastric cancer in a Chinese population. Gene. 2014;537(2):328–332. | ||
Livak KJ. Allelic discrimination using fluorogenic probes and the 5’ nuclease assay. Genet Anal. 1999;14(5–6):143–149. | ||
Abdulla MH, Valli-Mohammed MA, Al-Khayal K, et al. Cathepsin B expression in colorectal cancer in a Middle East population: potential value as a tumor biomarker for late disease stages. Oncol Rep. 2017; 37(6):3175–3180. | ||
Chen Y, Thai P, Wu R, et al. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. J Biol Chem. 2003;278(19):17036–17043. | ||
Park H, Li Z, Yang XO, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6(11):1133–1141 | ||
Yang XO, Pappu BP, Dong C, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28(1):29–39. | ||
Tosolini M, Kirilovsky A, Mlecnik B, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–1271. | ||
Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13(5):715–725. | ||
Ouyang W, Valdez P. IL-22 in mucosal immunity. Mucosal Immunol. 2008;1(5):335–338. | ||
Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13(2):139–145. | ||
Dai ZM, Zhang TS, Lin S, et al. Role of IL-17A rs2275913 and IL-17F rs763780 polymorphisms in risk of cancer development: an updated meta-analysis. Sci Rep. 2016;6:20439. | ||
Lu Y, Gu J, Lu H, et al. Association between IL-17A +197 G/A polymorphism and cancer risk: a meta-analysis. Genet Test Mol Biomarkers. 2016;20(1):24–30. | ||
Espinoza JL, Takami A, Nakata K, et al. A genetic variant in the IL-17 promoter is functionally associated with acute graft-versus-host disease after unrelated bone marrow transplantation. PLoS One. 2011;6(10):e26229. | ||
Dubin PJ, Kolls JK. Interleukin-17A and interleukin-17F: a tale of two cytokines. Immunity. 2009;30(1):9–11. | ||
Nemati K, Golmoghaddam H, Hosseini SV, et al. Interleukin-17FT7488 allele is associated with a decreased risk of colorectal cancer and tumor progression. Gene. 2015;561(1):88–94. | ||
Omrane I, Marrakchi R, Baroudi O, et al. Involvement of IL 17A, IL 17F and IL 23R polymorphism in colorectal cancer therapy. PLoS One. 2015;10(6):e0128911. | ||
Xu BL, Li YT, Dong SX, et al. IL-17 rs2275913 genetic variation contributes to the development of gastric cancer in a Chinese population. Genet Mol Res. 2016;15(2):1–7. | ||
Han L, Lee HS, Yoon JH, et al. Association of IL-17A and IL-17F single nucleotide polymorphisms with susceptibility to osteoarthritis in a Korean population. Gene. 20141;533(1):119–122. | ||
Hizawa N, Kawaguchi M, Huang SK, et al. Role of interleukin-17F in chronic inflammatory and allergic lung disease. Clin Exp Allergy. 2006; 36(9):1109–1114. | ||
Dai Z-M, Zhang T-S, Lin S, et al. Role of IL-17A rs2275913 and IL-17F rs763780 polymorphisms in risk of cancer development: an updated meta-analysis. Sci Rep. 2016; 6:20439. | ||
He Y, Du Y, Wei S, et al. IL-17A and IL-17F single nucleotide polymorphisms associated with lung cancer in Chinese population. Clin Respir J. 2017;11(2):230–242. | ||
Si FZ, Feng YQ, Han M. Association between interleukin-17 gene polymorphisms and the risk of laryngeal cancer in a Chinese population. Genet Mol Res. 2017;16(1):1–9. | ||
Arisawa T, Tahara T, Shiroeda H, et al. Genetic polymorphisms of IL17A and pri-microRNA-938, targeting IL17A 3′-UTR, influence susceptibility to gastric cancer. Hum Immunol. 2012;73(7):747–752. | ||
Grivennikov SI, Wang K, Mucida D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/Il-17-mediated tumor growth. Nature. 2012;491(7423):254–258. | ||
Petanidis S, Anestakis D, Argyraki M, et al. Differential expression of IL-17, 22 and 23 in the progression of colorectal cancer in patients with K-ras mutation: Ras signal inhibition and crosstalk with GM-CSF and IFN-γ. PLoS One. 2013;8(9):e73616. | ||
Wägsäter D, Löfgren S, Hugander A, et al. Expression of interleukin-17 in human colorectal cancer. Anticancer Res. 2006;26(6B):4213–4216. | ||
Bettelli E, Korn T, Oukka M, et al. Induction and effector functions of T(H)17 cells. Nature. 2008;453(7198):1051–1057. | ||
Langrish CL, Chen Y, Blumenschein WM, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201(2):233–240. | ||
Bjelica S, Santibanez JF. Novel patents targeting interleukin-17A: implications in cancer and inflammation. Recent Pat Anticancer Drug Discov. Epub 2018 Feb 19. | ||
Zhong W, Xu X, Zhu Z, et al. Increased interleukin-17A levels promote rituximab resistance by suppressing p53 expression and predict an unfavorable prognosis in patients with diffuse large B cell lymphoma. Int J Oncol. Epub 2018 Mar 5. | ||
Zhu Q, Zheng P, Chen X, et al. Andrographolide presents therapeutic effect on ulcerative colitis through the inhibition of IL-23/IL-17axis. Am J Transl Res. 2018;10(2):465–473. eCollection 2018. PMID: 29511440. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.