Back to Journals » Journal of Inflammation Research » Volume 10
IL-13 regulates human nasal epithelial cell differentiation via H3K4me3 modification
Authors Yu L, Li N , Zhang JS, Jiang Y
Received 15 August 2017
Accepted for publication 23 November 2017
Published 9 January 2018 Volume 2017:10 Pages 181—188
DOI https://doi.org/10.2147/JIR.S149156
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Lei Yu,1 Na Li,1 Jisheng Zhang,2 Yan Jiang1
1Department of Otorhinolaryngology, 2Key Laboratory of Otolaryngology-Head and Neck Surgery, Affiliated Hospital of Qingdao University, Qingdao, China
Introduction: Epigenetic regulation has been shown to play an important role in the development of inflammatory diseases, including chronic rhinosinusitis and nasal polyps. The latter are characterized by epithelial mis-differentiation and infiltration of inflammatory cytokines. H3K4me3 has been shown to be involved in regulating lineage commitment. However, the underlying mechanisms, especially in human nasal epithelial cells (HNEpC), remain underexplored. The objective of this study was to investigate the role of H3K4me3 in HNEpC differentiation treated with the Th2 cytokine IL-13.
Patients and methods: The expression levels of mRNA and proteins were investigated using reverse transcription-polymerase chain reaction (RT-PCR) assays and Western blot in nasal polyp tissues and human nasal epithelial cells respectively. We measured these levels of H3K4me3, MLL1 and targeted genes compared with control subjects.
Results: We demonstrate that expression of H3K4me3 and its methyltransferase MLL1 was significantly upregulated in IL-13-treated HNEpC. This elevation was also observed in nasal polyps. Expression of cilia-related transcription factors FOXJ1 and DNAI2 decreased, while goblet cell-derived genes CLCA1 and MUC5a increased upon IL-13 treatment. Mechanistically, knockdown of MLL1 restored expression of these four genes induced by IL-13.
Conclusion: These findings suggest that H3K4me3 is a critical regulator in control of nasal epithelial cell differentiation. MLL1 may be a potential therapeutic target for nasal inflammatory diseases.
Keywords: IL-13, H3K4me3 modification, nasal epithelial cell, differentiation
Introduction
The nasal epithelium possesses a pseudo-stratified structure, with specialized cell types including goblet cells, ciliated or non-ciliated columnar cells, and basal cells. This epithelium plays an important role in protecting the airway from infection, inflammation, and physical injury.1 Epithelial remodeling is induced upon damage, which is characterized by accumulation of pseudocyst formations, lack of collagen, and excessive inflammatory infiltrations, resulting in potentially irreversible structural changes.2 Epithelial cells undergo migration, proliferation, and differentiation in response to environment stimuli. The process is highly organized and is regulated by diverse growth factors and cytokines. Cytokines play essential roles in mediating allergic inflammation. Chronic rhinosinusitis (CRS) is a prevalent condition causing poor quality of life. CRS is divided into two subtypes: CRS with or without nasal polyps (CRSwNP or CRSsNP). CRSwNP displays epithelial barrier dysfunction with ciliary impairment and excessive mucus secretion.3 CRSwNP is characterized by a Th2 inflammatory pattern with high expression of type 2 cytokines IL-4, IL-5, and IL-13. CRSsNP merely expresses these biomarkers, but with high levels of INF-γ, characterized by predominance of Th1 cell.4–6 In addition, some CRS patients express a neutrophilic type of inflammation via Th1 and/or Th17 cells.7 Type 2 cytokines are induced by epithelium cells, which can activate type 2 innate lymphoid cells such as mast cells. These effector cells also produce type 2 cytokines, contributing to tissue remodeling. Nasal polyps display epithelial mucociliary dysfunction with excessive mucus secretion and cilia disappearance.8 Although the cause of nasal polyps is not clear, accumulated evidence suggests that cytokines play crucial roles. Previous studies suggest that IL-13 leads to stasis of sinonasal mucus production and cilia dysfunction,9 resulting in persistent inflammation. In human bronchial epithelial cells, IL-13 induced goblet cell hyperplasia and ciliated cell loss.10,11 The underlying mechanisms have yet to be fully explored.
Epigenetic regulation has been found to be involved in a number of inflammatory disorders.12–15 Previous studies16 have shown that, in nasal polyps, transcription and protein expression levels of HDCA2 are increased. Treatment with histone deacetylase inhibitor and histone modifications suppressed myofibroblast differentiation and altered extracellular matrix production.16,17 HDAC inhibitors suppressed inflammation via induction of FoxP3+ regulatory T-cells that also have relevance to asthma.18,19 In addition, in nasal polyp fibroblasts, H3 lysine27 acetylation (H3K27Ac) was highly expressed, suggesting that histone modifications regulated development of nasal polyps.20 It is noteworthy that increased active histone markers, including H3-K9 acetylation and H3-K4 trimethylation across the IL-4 and IFN-γ loci, were observed with Th1 or Th2 cell lineage commitment.21–23 It has been found that MLL1 can regulate the development of Th2 reactions by H3K4me3 modification through stabilizing expression of GATA3.24 MLL1 can also influence Th1 cell proliferation via regulating IL-12 responsiveness,25 suggesting that MLL1 has played a key role of regulating cellular inflammatory processes.
In the present study, we observed elevated H3K4me3 expression in the nasal polyps. We investigated the function of the histone methyltransferase MLL1 in human nasal epithelial cells (HNEpC) upon IL-13 treatment. We found that H3K4me3 may play an important role in the mis-differentiation of nasal epithelium in inflammatory disorders.
Patients and methods
Subject collection
Nasal polyp tissues and normal inferior turbinate tissues were collected from 16 patients with CRSwNP undergoing functional endoscopic sinus surgery from the Department of Otorhinolaryngology, Affiliated Hospital of Qingdao University, China. All tissues were used immediately and/or snap-frozen at -80°C. The study was approved by the local ethics committee and the regulatory authorities of China. Written informed consent was obtained from all subjects before sample collection. Patients with an established immunodeficiency and pregnancy were excluded from the study. None of the patients had allergy, asthma, or aspirin sensitivity and treated with corticosteroids for at least 1 month before surgery.
Cell culture and treatment with IL-13
HNEpCs were purchased from PromoCell (Atlanta, GA, USA) and cultured in 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (Thermo Fisher Scientific) in 5% CO2 atmosphere at 37°C. The cells were stimulated with Recombinant Human IL-13 (2#00-13, Escherichia coli; Peprotech, Rocky Hill, NJ, USA) at a final concentration of 5, 10, 50, 100, and 200 ng/mL for indicated time.
Plasmids and transfection
MLL1 shRNA plasmids were obtained from GeneChem Company (Montreal, QC, Canada). The targeted sequences were 5′-GATTATGACCCTCCAATTAAA-3′ and 5′-GCACTGTTAAACATTCCACTT-3′. The plasmids were transfected with helper vectors, pDelta8.9 and pVSV-G, into HEK293FT cells. After 48 h of transfection, the medium was collected and centrifuged at 50,000 ×g for 3 h. The pellets were resuspended in PBS. Lentivirus was transduced into the HNEpC. To obtain a stable and pure MLL1-knockdown cell population, we performed selection with 2 μg/mL of puromycin after 48 h of transfection. It usually takes 2 days for all the control cells to die. After selection, we collected cells and examined the efficiency of transfection through real-time polymerase chain reaction (PCR) and Western blot.
Western blot analysis
To obtain cell and tissue proteins, samples were processed with 2% sodium dodecyl sulfate (SDS) lysis buffer and sonicated to break up DNA. Lysates were boiled for 10 min at 98°C. Then, the samples were measured by BCA Protein Assay Kit (Beyotime Institute of Biotechnology, Shanghai, China), and 20 μg of total protein was loaded. Transferred polyvinylidene fluoride membranes were incubated with primary antibodies against H3K4me3 (1:1000; #GC-263, PTMbiolabs, Chicago, IL, USA), MLL1 (1:1000; #14197, Cell Signaling Technology, Danvers, MA, USA), and β-tubulin (1:3000; Beyotime Institute of Biotechnology) overnight at 4°C, followed by incubation with secondary antibodies of anti-mouse IgG and anti-rabbit IgG, respectively (1:2000; CWbiotech, Beijing, China) for 1 h at room temperature. Western blot analyses were normalized to β-tubulin. The blots were developed with Super Signal Pico substrate (Pierce Biotechnology, Shanghai, China). Each immunoblot was repeated three times, with samples obtained from different experiments. The relative intensity of protein bands was measured with NIH image J software.
RNA preparation and quantitative real-time qPCR
Samples were stored at -80°C until homogenization and no more than 25 mg tissues were homogenized in Trizol. For quantitative real-time PCR, total RNA was extracted from HNEpC and tissues using RNAiso Plus (D9108; Takara Bio, Tokyo, Japan) following the instructions from the manufacturer. RNA quantity and purity were determined by Nanodrop spectrophotometer. GAPDH was used as an internal control. Reverse cloning of cDNA by 500 ng RNA was performed using a First Strand cDNA Synthesis Kit (RR037A; Takara) according to the manufacturer’s instructions. Real-time PCR was performed to determine the mRNA expression. In brief, real-time PCR was conducted using the Roche Lightcycler480 Real-time PCR System with SYBR green reagents from Takara (RR820A). Quantifications were normalized to GAPDH. Relative gene expression was calculated using the 2-ΔΔCt method.
The primer sequences used for application were as follows:
FOXJ1 forward: 5′-GTTCTCCCGAGGCACTTTGA-3′ and reverse: 5′-CACCAAGATCACCCTGTCGG-3′; DNAI2: forward: 5′-GTTGAGGGTCAAGAGGTGGG-3′ and reverse: 5′-GGATGAGGAGCACCGATG-3′; MUC5a: forward: 5′-ACCCATGGAATTCGGGAACC-3′ and reverse: 5′-TTGATCACCACCGTCTG-3′; CLCA1: forward: 5′-TGGTAACCGCCTCAATCGAC-3′ and reverse: 5′-GCCAACCTTAGCAATGCCTG-3′; GAPDH: forward: 5′-TCGACAGTCAGCCGCATCTT-3′ and reverse: 5′-GAGTTAAAAGCAGCCCTGGTG-3′; MLL1: forward: 5′-TTTAGAGGAGAACGAGCGCC-3′ and reverse: 5′-AGGGTGATAGCTGTTTCGGC-3′; MLL2: forward: 5′-GTCGCAAGCATAAGACGACC-3′ and reverse: 5′-ACCATCCGTTCTGTGCCTTC-3′; MLL3: forward: 5′-TCCTCGGCTCCAACAAAATCT-3′ and reverse: 5′-CAGGACCAATATCTGAATGATCAAC-3′; MLL4: forward: 5′-AAACGGCCCCATACCCTGA-3′ and reverse: 5′-GTTGTTCTTCCATTCGGTGCG-3′; MLL5: forward: 5′-GCCATTTTCCCAGAGCGAGA-3′ and reverse: 5′-TGTCTATGCCCACTCTGTTGC-3′; SETD1A: forward: 5′-CGTTGCCATGTCAGGTCCAA-3′ and reverse: 5′-GCACGTTGTCATTCAGCCTT-3′; and JARID1B: forward: 5′-CATATCTGCCCAATGGTGCG-3′ and reverse: 5′-TCTAACACTGGCACACGTCC-3′.
Statistical analyses
Statistical analyses were done using descriptive and inferential statistics by GraphPad Prism software version 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t-test and ANOVA test were performed to determine the statistical significance between two groups and among more than two groups, respectively. All in vitro experiments were done and repeated at least three times. In Figures 1–3, mean value ± 1 SD are presented. For all statistics, P=0.05 was considered to be statistically significant.
Results
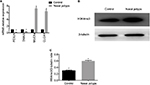
Increased expression of H3K4me3 and relative epithelial gene mRNA expression in nasal polyps
Pathological remodeling of nasal polyps is characterized by epithelial dysfunction. First, we collected nasal polyp tissues and inferior turbinate samples from the same side of nasal polyps patients undergoing polypectomy for the treatment of nasal obstruction. mRNA expression of FOXJ1, DNAI2, CLCA1, and MUC5a was examined. Expression of FOXJ1 and DNAI2, major cilia-related transcription factors, was decreased in nasal polyps compared to control, whereas that of CLCA1 and MUC5a, goblet cell-derived genes, was elevated (Figure 1A), suggesting mis-differentiation of epithelium. H3K4me3 expression was increased in nasal polyps samples compared with control (Figure 1B and 1C), suggesting that histone methylation may play an important role in metaplasia of nasal epithelia.
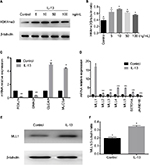
Elevation of H3K4me3 and MLL1 expression upon IL-13 treatment in HNEpC
To further understand the role of H3K4me3 in progression of nasal Th2 inflammatory diseases, we treated HNEpC cells with IL-13 at varying concentrations. H3K4me3 expression was elevated by IL-13 treatment (Figure 2A). Peak expression of H3K4me3 occurred at 10 ng/mL concentration of IL-13 (Figure 2B). Thus, we chose 10 ng/mL concentration of IL-13 for further experiments. Next, we analyzed mRNA expression of FOXJ1, DNAI2, CLCA1, and MUC5a. IL-13 induced mRNA expression of CLCA1 and MUC5a, but suppressed FOXJ1 and DNAI2 in HNEpCs (Figure 2C). This is consistent with the results obtained for nasal polyps tissues.
H3K4me3 is a dynamic and reversible process that is governed by histone methyltransferases and demethylases. We examined a variety of important genes regulating the methyl group of H3K4me3 by quantitative real-time PCR. The methyltransferase genes (MLL1, MLL2, MLL3, MLL4, MLL5, and SETD1A) were elevated and demethylase Jarid1b was decreased with IL-13 treatment (Figure 2D). Of these, MLL1 showed the greatest change. We validated IL-13-induced elevation of MLL1 by Western blot (Figure 2E). These results suggest that IL-13-induced H3K4me3 elevation was probably regulated by both increase in methyltransferase and reduction of demethylase. However, the methyltransferase MLL1 plays a predominant role.
Knockdown of MLL1 reversed IL-13-induced changes of gene expression
To evaluate whether MLL1 influences IL-13-induced metaplasia, we knocked down MLL1 with lentivirus and examined mRNA expression of FOXJ1, DNAI2, MUC5a, and CLCA1. First, we validated MLL1 shRNA efficiency. Compared with control, MLL1 mRNA and protein expression was significantly reduced upon MLL1 knockdown (Figure 3A and B), and this was proved (Figure 2F). As expected, H3K4me3 expression markedly declined with MLL1 reduction (Figure 3B). The expression of FOXJ1 and DNAI2 significantly increased, while that of CLCA1 and MUC5a decreased compared with control (Figure 3C). These data suggest that MLL1 knockdown reverses the alterations of hallmark genes in nasal epithelium induced by inflammation.
Discussion
To our knowledge, this study is the first to investigate the connection between H3K4me3 and HNEpC metaplasia by IL-13. We provide additional evidence in favor of histone methylation involvement in nasal inflammation.
Histone modifications and DNA methylation are crucial for sustaining distinct gene expression.26 Cho et al16 showed that in nasal polyps, expression of HDAC2 increased compared to normal nasal inferior turbinates. We found that expression of H3K4me3 increased in nasal polyps compared with controls.
Many studies have suggested that a variety of Th2 inflammatory cytokines are implicated in development and maintenance of nasal inflammation, such as three representative cytokines (IL-4, IL-5, and IL-13).27,28 It is widely accepted that increased IL-4 and IL-13 can upregulate eotaxin production in epithelial cells, and IL-5 plays pivotal role in the recruitment and survival of eosinophils,29 which is also positively associated with serum total IgE.30 Nasal polyp tissue significantly highly expressed IL-5 and IgE in CRSwNP compared with CRSsNP patients,7 which may lead to nasal polyp formation at specific sites with mucosal inflammation. As for immunoglobulin production, total IgE has also often been highly expressed within polyp tissue and fluid, especially in eosinophil CRSwNP, which in turn can contribute to local inflammation.31–33
H3K4 methylation has played an important role in regulating inflammatory gene transcription through different ways. For example, Li et al34 found that SET7/9, H3K4 methyltransferase, was involved in gene expression of TNF-α via recruitment of NF-κB p65 to inflammatory gene promoters in inflammation and immunity. Enrichment of H3K4me3 could increase the expression of IFN-γ and IL-4 produced by Th17 cells through interrupting the balance between native CD4+ T-cell precursors and Th1, Th2, and Th17 T-helper cell subsets in asthma,35,36 and IL-4 stimulation could increase H3K4me3 at IgE locus in CL-01 and primary B cells, resulting in high expression of IgE.37,38
IL-13 has been widely recognized as an important cytokine in this process.39–41 In our study, we found that expression of H3K4me3 and MLL1 showed higher levels in IL-13-induced HNEpCs. To our knowledge, this is the first study to show that H3K4me3 and MLL1 expression levels increased in nasal polyps. As for the status of H3K4me3 in CRSwNP, further studies should be applied according to diverse clusters, which would be informative for individual treatment.
Increased gene expression of CLCA1 and MUC5AC and eosinophilic infiltration is seen following instillation of IL-13 into mouse airway,42 mediated by the JAK/STAT6 pathway.43 In nasal polyps, STAT6-positive cells were localized in epithelium, gland cells, and inflammatory cells, and expression of STAT6 in epithelium had significantly increased compared with the control, which was positively associated with the recruitment of eosinophils.44 In the airway epithelium, IL-13 treatment decreased FOXJ1 mRNA expression via binding of STAT6 to the FOXJ1 promoter.45,46 We found increased MUC5A and CLCA1 and decreased FOXJ1 and DNAI2 at the transcriptional level with IL-13 treatment in HNEpC, consistent with previous reports.58 Recently, H3K4me3, an activated histone marker, was shown to be important in inflammatory processes,47 especially with high expression of IL-13.48 Our results suggest that H3K4me3 expression increases upon IL-13 treatment, which is at least partially attributed to methyltransferase MLL1. We examined four genes following MLL1 knockdown. It appears that CLCA1 and MUC5A expression is positively regulated by MLL1, although H3K4me3 enrichment at the promoter of these two genes needs to be further investigated. Carson et al49 found that MLL1-dependent H3K4me3 modification could regulate macrophage proinflammatory responses, indicating that MLL1 could be a novel therapeutic target for inflammatory diseases. Regarding FOXJ1 and DNAI2, the underlying mechanism remains to be explored in future. The signaling pathway of H3K4me3 and MLL1 remains poorly understood. Some studies have found that histone acetylation regulates chronic inflammatory disorders induced by IL-13 via STAT6 signaling pathway.50 We suspect that H3K4me3 may be involved in the STAT6 pathway. In addition, some studies have shown that Th2 cytokines such as IL-13 induce the airway inflammatory environment via the mitogen-activated protein kinase (MAPK) pathway.51,52 Histone modification could affect the p38 MAPK pathway, suggesting that histone modifications affect inflammatory development induced by IL-13 through MAPK.53,54 The function of the H3K4me3 and MLL1 signaling network needs to be further investigated.
Meanwhile, some studies have forced the necessitiy to develop inhibitors of MLL1 methyltransferase activity. MM-401 was able to inhibit MLL1 activity by blocking MLL1–WDR5 interaction without affecting other MLL family histone methyltransferases,55–57 supporting that MLL1 could be the target of epigenetic therapy. MLL1 may be a potential target for CRSwNP via influencing the process of H3K4me3 modification, especially in Th2 cytokine-dominant patients.
Disclosure
The authors report no conflicts of interest in this work.
References
Yan Y, Gordon WM, Wang DY. Nasal epithelial repair and remodeling in physical injury, infection, and inflammatory diseases. Curr Opin Otolaryngol Head Neck Surg. 2013;21(3):263–270. | ||
Bachert C, Pawankar R, Zhang L, et al. ICON: chronic rhinosinusitis. World Allergy Organ J. 2014;7(1):25. | ||
Yeh TH, Lee SY, Hsu WC. Expression of SPLUNC1 protein in nasal polyp epithelial cells in air-liquid interface culture treated with IL-13. Am J Rhinol Allergy. 2010;24(1):17–20. | ||
Van Zele T, Claeys S, Gevaert P, et al. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006;61(11):1280–1289. | ||
Pietruszewska W, Olejniczak I, Durko T, Mlynarski W. Udział IFN-gamma i TNF-alfa w etiopatogenezie polipów nosa - badania wstępne. [Role of IFN-gamma and TNF-alpha in etiology of nasal polyps – initial studies]. Otolaryngol Pol. 2008;62(1):54–58. Polish. | ||
Milonski J, Zielinska-Blizniewska H, Majsterek I, et al. Expression of POSTN, IL-4, and IL-13 in chronic rhinosinusitis with nasal polyps. DNA Cell Biol. 2015;34(5):342–349. | ||
Shi LL, Xiong P, Zhang L, et al. Features of airway remodeling in different types of Chinese chronic rhinosinusitis are associated with inflammation patterns. Allergy. 2013;68(1):101–109. | ||
Seong JK, Koo JS, Lee WJ, et al. Upregulation of MUC8 and downregulation of MUC5AC by inflammatory mediators in human nasal polyps and cultured nasal epithelium. Acta Otolaryngol. 2002;122(4):401–407. | ||
Antunes MB, Gudis DA, Cohen NA. Epithelium, cilia, and mucus: their importance in chronic rhinosinusitis. Immunol Allergy Clin North Am. 2009;29(4):631–643. | ||
Grosse-Onnebrink J, Werner C, Loges NT, et al. Effect of TH2 cytokines and interferon gamma on beat frequency of human respiratory cilia. Pediatr Res. 2016;79(5):731–735. | ||
Sun L, Ren X, Wang IC, et al. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci Signal. 2017;10(475):eaai8583. | ||
Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. J Cell Mol Med. 2010;14(6A):1225–1240. | ||
Pearce EL, Shen H. Making sense of inflammation, epigenetics, and memory CD8+ T-cell differentiation in the context of infection. Immunol Rev. 2006;211:197–202. | ||
Adcock IM. HDAC inhibitors as anti-inflammatory agents. Br J Pharmacol. 2007;150(7):829–831. | ||
Stockley RA, Mannino D, Barnes PJ. Burden and pathogenesis of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6(6):524–526. | ||
Cho JS, Moon YM, Park IH, et al. Epigenetic regulation of myofibroblast differentiation and extracellular matrix production in nasal polyp-derived fibroblasts. Clin Exp Allergy. 2012;42(6):872–882. | ||
Cho JS, Moon YM, Park IH, et al. Effects of histone deacetylase inhibitor on extracellular matrix production in human nasal polyp organ cultures. Am J Rhinol Allergy. 2013;27(1):18–23. | ||
Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. | ||
Wang L, Tao R, Hancock WW. Using histone deacetylase inhibitors to enhance Foxp3(+) regulatory T-cell function and induce allograft tolerance. Immunol Cell Biol. 2009;87(3):195–202. | ||
Cahill KN, Raby BA, Zhou X, et al. Prostanoid2 expression and resistance to prostaglandin E2 in nasal polyp fibroblasts from subjects with aspirin-exacerbated respiratory disease. Am J Respir Cell Mol Biol. 2016;54(1):34–40. | ||
Kumar RK, Hitchins MP, Foster PS. Epigenetic changes in childhood asthma. Dis Model Mech. 2009;2(11–12):549–553. | ||
Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16(5):649–660. | ||
Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30(1):155–167. | ||
Yamashita M, Hirahara K, Shinnakasu R, et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24(5):611–622. | ||
Schaller M, Ito T, Allen RM, et al. Epigenetic regulation of IL-12-dependent T cell proliferation. J Leukoc Biol. 2015;98(4):601–613. | ||
Ausio J, Levin DB, De Amorim GV, Bakker S, Macleod PM. Syndromes of disordered chromatin remodeling. Clin Genet. 2003;64(2):83–95. | ||
Bachert C, Wagenmann M, Hauser U, Rudack C. IL-5 synthesis is upregulated in human nasal polyp tissue. J Allergy Clin Immunol. 1997;99(6 pt 1):837–842. | ||
Min YG, Lee CH, Rhee CS, Hong SK, Kwon SH. Increased expression of IL-4, IL-5, IFN-gamma, IL-6, IL-8, and TGF-beta mRNAs in maxillary mucosa of patients with chronic sinusitis. Am J Rhinol. 1999;13(5):339–343. | ||
Otto BA, Wenzel SE. The role of cytokines in chronic rhinosinusitis with nasal polyps. Curr Opin Otolaryngol Head Neck Surg. 2008;16(3):270–274. | ||
Tomassen P, Vandeplas G, Van Zele T, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016;137(5):1449–1456.e4. | ||
Perez-Novo CA, Kowalski ML, Kuna P, et al. Aspirin sensitivity and IgE antibodies to Staphylococcus aureus enterotoxins in nasal polyposis: studies on the relationship. Int Arch Allergy Immunol. 2004;133(3):255–260. | ||
Van Zele T, Gevaert P, Watelet JB, et al. Staphylococcus aureus colonization and IgE antibody formation to enterotoxins is increased in nasal polyposis. J Allergy Clin Immunol. 2004;114(4):981–983. | ||
Van Zele T, Gevaert P, Holtappels G, van Cauwenberge P, Bachert C. Local immunoglobulin production in nasal polyposis is modulated by superantigens. Clin Exp Allergy. 2007;37(12):1840–1847. | ||
Li Y, Reddy MA, Miao F, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283(39):26771–26781. | ||
Rowell E, Wilson CB. Programming perpetual T helper cell plasticity. Immunity. 2009;30(1):7–9. | ||
Singh A, Yamamoto M, Ruan J, et al. Th17/Treg ratio derived using DNA methylation analysis is associated with the late phase asthmatic response. Allergy Asthma Clin Immunol. 2014;10(1):32. | ||
Fear DJ, McCloskey N, O’Connor B, Felsenfeld G, Gould HJ. Transcription of Ig germline genes in single human B cells and the role of cytokines in isotype determination. J Immunol. 2004;173(7):4529–4538. | ||
Dayal S, Nedbal J, Hobson P, et al. High resolution analysis of the chromatin landscape of the IgE switch region in human B cells. PLoS One. 2011;6(9):e24571. | ||
Mulligan JK, Mulligan RM, Atkinson C, Schlosser RJ. Human sinonasal epithelial cells direct dendritic function and T-cell T helper 1/T helper 2 skewing following Aspergillus exposure. Int Forum Allergy Rhinol. 2011;1(4):268–274. | ||
Woodworth BA, Joseph K, Kaplan AP, Schlosser RJ. Alterations in eotaxin, monocyte chemoattractant protein-4, interleukin-5, and interleukin-13 after systemic steroid treatment for nasal polyps. Otolaryngol Head Neck Surg. 2004;131(5):585–589. | ||
de Castro MC, Rocha-Silva F, Gomes LI, et al. Impact of mitomycin C on the mRNA expression signatures of immunological biomarkers in eosinophilic nasal polyposis. Am J Rhinol Allergy. 2013;27(1):e32–e41. | ||
Mertens TC, Hiemstra PS, Taube C. Azithromycin differentially affects the IL-13-induced expression profile in human bronchial epithelial cells. Pulm Pharmacol Ther. 2016;39:14–20. | ||
Nakano T, Inoue H, Fukuyama S, et al. Niflumic acid suppresses interleukin-13-induced asthma phenotypes. Am J Respir Crit Care Med. 2006;173(11):1216–1221. | ||
Cao Q, Zhang T, Wang L, Luo S, Tu Z. [Expression of STAT6 in human nasal polyps and the relation between STAT6 and eosinophil infiltration]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2009;23(20):917–919. Chinese. | ||
Gomperts BN, Kim LJ, Flaherty SA, Hackett BP. IL-13 regulates cilia loss and foxj1 expression in human airway epithelium. Am J Respir Cell Mol Biol. 2007;37(3):339–346. | ||
Wu CA, Peluso JJ, Shanley JD, Puddington L, Thrall RS. Murine cytomegalovirus influences Foxj1 expression, ciliogenesis, and mucus plugging in mice with allergic airway disease. Am J Pathol. 2008;172(3):714–724. | ||
Zhang J, An X, Han Y, et al. Overexpression of JARID1B promotes differentiation via SHIP1/AKT signaling in human hypopharyngeal squamous cell carcinoma. Cell Death Dis. 2016;7(9):e2358. | ||
Yashiro T, Kubo M, Ogawa H, Okumura K, Nishiyama C. PU.1 suppresses Th2 cytokine expression via silencing of GATA3 transcription in dendritic cells. PLoS One. 2015;10(9):e0137699. | ||
Carson WF, 4th, Cavassani KA, Soares EM, et al. The STAT4/MLL1 epigenetic axis regulates the antimicrobial functions of murine macrophages. J Immunol. 2017;199(5):1865–1874. | ||
Lim EJ, Lu TX, Blanchard C, Rothenberg ME. Epigenetic regulation of the IL-13-induced human eotaxin-3 gene by CREB-binding protein-mediated histone 3 acetylation. J Biol Chem. 2011;286(15):13193–13204. | ||
Ma Y, Zhang JX, Liu YN, et al. Caffeic acid phenethyl ester alleviates asthma by regulating the airway microenvironment via the ROS-responsive MAPK/Akt pathway. Free Radic Biol Med. 2016;101:163–175. | ||
Wang B, Gao Y, Zheng G, et al. Platycodin D inhibits interleukin-13-induced the expression of inflammatory cytokines and mucus in nasal epithelial cells. Biomed Pharmacother. 2016;84:1108–1112. | ||
Kikuchi H, Yuan B, Yuhara E, Takagi N, Toyoda H. Involvement of histone H3 phosphorylation through p38 MAPK pathway activation in casticin-induced cytocidal effects against the human promyelocytic cell line HL-60. Int J Oncol. 2013;43(6):2046–2056. | ||
Kikuchi H, Yuan B, Yuhara E, et al. Involvement of histone H3 phosphorylation via the activation of p38 MAPK pathway and intracellular redox status in cytotoxicity of HL-60 cells induced by Vitex agnus-castus fruit extract. Int J Oncol. 2014;45(2):843–852. | ||
Karatas H, Townsend EC, Cao F, et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc. 2013;135(2):669–682. | ||
Senisterra G, Wu H, Allali-Hassani A, et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J. 2013;449(1):151–159. | ||
Cao F, Townsend EC, Karatas H, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell. 2014;53(2):247–261. | ||
Jiao J, Duan S, Meng N, Li Y, Fan E, Zhang L. Role of IFN-gamma, IL-13, and IL-17 on mucociliary differentiation of nasal epithelial cells in chronic rhinosinusitis with nasal polyps. Clin Exp Allergy. 2016;46(3):449–460. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.