Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 17
IFN-γ, SCF, MIP1b and IL-16 Were Associated with Risk of Diabetic Nephropathy: A Mendelian Randomization Study
Authors An L, Ren X, Pan Y, Gao W, Ren L, Wang J , Wang Y
Received 28 November 2023
Accepted for publication 3 February 2024
Published 22 February 2024 Volume 2024:17 Pages 851—856
DOI https://doi.org/10.2147/DMSO.S452227
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Juei-Tang Cheng
Li An,1,2,* Xiaomei Ren,1,* Ye Pan,2 Wei Gao,1 Liqun Ren,1 Jing Wang,3 Yao Wang2
1Department of Geriatrics, ZhongDa Hospital, Southeast University School of Medicine, Nanjing, 210009, People’s Republic of China; 2Department of Endocrine, ZhongDa Hospital, Southeast University School of Medicine, Nanjing, 210009, People’s Republic of China; 3Yizheng Hospital of Nanjing Drum Tower Hospital Group, Yizheng, 211400, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jing Wang; Yao Wang, Tel/Fax +86-25- 83272111, Email [email protected]; [email protected]
Background: The impact of inflammatory factors on the risk of diabetic nephropathy (DN) is inconsistent. Two-sample Mendelian randomization (MR) analyses were used to detect the causal role of inflammatory factors in DN risk.
Methods: Inflammatory factor GWAS summary data were collected from a meta-analysis including 8,293 Finnish participants, and DN information was extracted from a GWAS of 213,746 individuals from FinnGen. The MR Pleiotropy Residual Sum and Outlier (MR-PRESSO) outlier test was used for the removal of horizontal pleiotropic outliers. Multivariable MR analysis was also used to adjust for pleiotropy.
Results: IFN-γ [ORIVW: 1.33; 95% CI: 1.09– 1.63; p=0.005] and SCF [ORIVW: 1.25, 1.02– 1.52; p = 0.027] were associated with an increased risk of DN. MIP1b [ORIVW: 0.92; 95% CI: 0.85– 0.98; p = 0.022] and IL-16 [ORIVW: 0.89, 0.81– 0.99; p = 0.043] showed negative associations with the risk of DN. We validated our MR results with MR-PRESSO analyses. Significant horizontal pleiotropy was not found. Moreover, in the multivariable MR analysis, the associations between cytokines and DN risk remained.
Conclusion: Our MR results based on genetic data contribute to a better understanding of the pathogenesis of DN and provide evidence for a causal effect of inflammatory factors on DN. These findings support targeting specific inflammatory factors to alleviate DN risk.
Keywords: causal association, diabetic nephropathy, inflammatory factors, Mendelian randomization
Introduction
Diabetes-related complications are considered one of the most challenging health problems worldwide. Diabetic nephropathy (DN) is one of the most frequent and severe microvascular complications of diabetes mellitus, which develops in approximately one-third of patients with diabetes and is a more frequent primary cause of end-stage renal failure.1 Risk factors for DN are classified into modifiable and nonmodifiable factors. The modifiable factors include obesity, hypertension, hyperglycemia, dyslipidemia,2 and smoking.3 The second group comprises age, sex, ethnicity,4 and genetic variants (CARS, FRMD3, viz. NLRP3, INPPL1, PIK3C2G, NRXN3, and TBC1D4).5
Inflammatory molecules play a key role in glomerular and tubulointerstitial damage associated with urinary albumin excretion (UAE) among patients with diabetes.6,7 These molecules can also recruit inflammatory cells (monocytes, neutrophils, and lymphocytes) to accumulate in the kidney, which is associated with the development and progression of DN.8,9 There are no available treatments to prevent the development of DN. Assessing the causal role of inflammatory factors in DN risk may help in protecting patients from the development and progression of renal damage.
The existence of a potential causal association between exposure and outcome can be estimated by using the Mendelian randomization (MR) approach with genetic variants.10,11 We used two-sample MR analyses to explore the potential causal associations between inflammatory factors and the risk of DN. Our findings will help identify the important role of inflammatory factors in the development and progression of DN and provide potential implications for the pathogenesis, diagnosis and therapy of DN.
Methods
Study Design
The present study followed the Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization (STROBE-MR) guidelines. A brief description of the study design is displayed in Figure 1.
Data Source
The GWAS summary data for 41 inflammatory factors were obtained from a meta-analysis on three independent cohorts (YFS, FINRISK1997 and FINRISK2002), including 8,293 Finnish participants (Supplementary Table 1). DN data were extracted from a GWAS of 213,746 individuals of European ancestry from FinnGen (3,282 cases vs 210,463 controls). All data were downloaded from the MRC Integrated Epidemiology Unit (IEU) open GWAS database (https://gwas.mrcieu.ac.uk/datasets/).12
Instrumental Variables (IVs)
Since only a few independent genetic variants reached genome-wide significance (p <5×10−8), we employed a less stringent threshold (5×10−6) to obtain more SNPs for inflammatory factors. We clumped these SNPs with linkage disequilibrium (LD) r2 < 0.01. We deleted weak IVs with an F value less than 10.13 The MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) test was used for the removal of horizontal pleiotropic outliers.14 SNPs missing in the outcome were replaced by proxies with LD R2 >0.8.
Statistical Analysis
The inverse variance weighted (IVW) method was administered as the major statistical method, which is to combine the specific ratio estimates using each SNP.15 The IVW method uses an asymptotic estimation of the standard error of causal ratio for each variable; this is known to underestimate the true changes in estimation, especially when asymptotic estimation is weak.16 MR-PRESSO was also performed to validate the IVW results. MR results were reported as odds ratios (ORs) with 95% confidence intervals (CIs). The statistical power of MR was calculated using an online website (sb452.shinyapps.io/power/). MR-Steiger analysis was used to monitor the direction of the potential causal effect between inflammatory factors and DN risk. Cochran’s Q-test was performed to detect heterogeneity between IVs. If heterogeneity existed, the IVW random-effect model was used. The MR-PRESSO global test and pleiotropy test function of the TwoSampleMR package were used to detect horizontal pleiotropy. The robustness of the MR analysis was assessed by leave-one-out analysis. TwoSampleMR and MR-PRESSO packages were used for statistical tests. A p-values below 0.0012 (0.05/41) was considered as strong evidence of associations using Bonferroni method to correct for multiple testing. We also selected SNPs for positive associations to perform multivariable MR analyses to adjust for pleiotropy. A p value of 0.05 was the threshold to indicate statistical significance.
Results
As shown in Figure 2, IFN-γ [ORIVW: 1.33; 95% CI: 1.09–1.63; p= 0.005] and SCF [ORIVW: 1.25, 1.02–1.52; p =0.027] showed a causal relationship with an increased risk of DN. MIP1b [ORIVW: 0.92; 95% CI: 0.85–0.98; p = 0.022] and IL-16 [ORIVW: 0.89, 0.81–0.99; p = 0.043] showed a causal relationship with a decreased risk of DN. There was a strong trend toward a positive association between SDF1a and the risk of DN [ORIVW: 1.26, 0.99–1.59; p = 0.051]. The same associations were observed using the MR-PRESSO method (Supplementary Table 2). Neither IVW nor MR-PRESSO found any significance for multiple testing. Significant horizontal pleiotropy was not found in our study. In multivariable IVW analyses, We also observe the same associations (SCF [ORIVW: 1.32, 1.10–1.59; p = 0.003]; IL-16 [ORIVW: 0.90, 0.82–0.99; p = 0.031]; MIP1b [ORIVW: 0.92; 95% CI: 0.86–0.98; p = 0.010]; IFN-γ [ORIVW: 1.20; 95% CI: 1.01–1.43; p =0.043]).
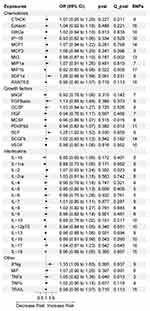 |
Figure 2 Associations between genetically predicted inflammatory factors and DN risk. |
Discussion
In this study, we conducted an MR analysis to assess the causal association of inflammatory factors with DN. We successfully identified four upstream regulators of DN. Higher IFN-γ and SCF were associated with an increased risk of DN, while higher MIP1b and IL-16 were shown to possibly decrease the risk of DN. Our results confirmed the hypothesis that inflammatory factors play an important role in the development of DN.
First, we explicate the surprisingly positive causality between IFN-γ and DN. IFN-γ secretion is considered to be a powerful upstream event for many inflammatory responses and a major mediator for the activation or release of other inflammatory cytokines, such as TNF-α and the IL-1 families.17 Elevated serum levels of IFN-γ have been observed in type-2 diabetes patients with nephropathic complications compared with those of healthy controls.18,19 IFN-γ mRNA expression was significantly increased in diabetic mouse kidneys compared with those of controls.20 Second, SCF is a novel endothelial permeability factor.21 SCF/c-kit signaling promotes the recruitment of endothelial progenitor cells and contributes to neovascularization.22,23 Neovascularization has been implicated in the genesis of diverse diabetic complications, such as retinopathy and nephropathy.24 Increased expression of SCF protein and mRNA in DN model rats was positively correlated with the infiltration degree of mast cells, which may aggravate renal tubulointerstitial fibrosis in DN rats.25 Third, IL-16 may contribute to regulatory T-cell (Treg) expansion for immunosuppressive effects.26 In T1D, peripheral immunocompetent cells are defective in IL-16 secretion.27 Lower expression of CD4+CD25+Foxp3+ Tregs was associated with microalbuminuria and macroalbuminuria in type-2 diabetes patients.28 Adoptive transfer of CD4+FoxP3+ Tregs significantly improved diabetic nephropathy by limiting the proinflammatory milieu.29 Fourth, MIP-1b is a chemokine also known as CCL4. MIP-1b plays an important role in the immunoregulatory process, recruiting Tregs into inflammatory sites.30 Tregs deficient in CCL4 expression were impaired in their ability to suppress experimental autoimmune encephalomyelitis in murine models.31 CCL4 was significantly lower in patients with type-1 diabetes mellitus (DM) than it was in control participants.32 A protective role of CCL4 was suggested in a nonobese diabetic (NOD) mouse model.33
There are a few study limitations that should be considered. First, our study was based on the available GWAS data. There are no data to infer the differential effects of sex on the causal association of inflammatory factors with DN. Sexual dimorphism may play a key role in the pathogenesis of DN.34 Second, the study population was limited to individuals of European ethnicity. Our results cannot be generalized to other populations. Race/ethnicity is classified as a susceptibility risk factor for DN.1 Third, MR results could reflect the lifelong effect of inflammatory factors on DN. However, it may be directly affected in adult life due to many unknown factors. Fourth, we only obtained GWAS data 41 inflammatory factors from a meta-analyses involving 8293 individuals35 and some essential inflammatory factors are not included in our study. TGF-β/Smad signaling plays a critical role in the development of DN.36 Finally, our results were based on the reported GWAS summary data noted above. Using other data may yield different results.
Conclusion
In conclusion, this MR study suggested that there was a causal effect of inflammatory factors on the risk of DN. Four common inflammatory factors, IFN-γ, SCF, MIP1b and IL-16, should be noted and emphasized in the pathogenesis and treatment of DN. Further study is needed to validate our findings in a prospective manner.
Data Sharing Statement
Only publicly available data were used in our study, and data sources are described in the Methods/Supplementary material.
Ethics Statement
Our study was based on publicly available GWAS summary-level data and all original studies have been approved by their Institutional Review Boards or local ethics committees. Besides, our study did not involve individual-level data and was deemed not to require ethical approval after consultation with the Ethics Committee of ZhongDa Hospital.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Rz A, Mt R, Kr T. Diabetic kidney disease: challenges, progress, and possibilities. Clin J Am Soc Nephrol. 2017;12(12):2032–2045. doi:10.2215/CJN.11491116
2. Amatruda M, Gembillo G, Giuffrida AE, Santoro D, Conti G. The aggressive diabetic kidney disease in youth-onset type 2 diabetes: pathogenetic mechanisms and potential therapies. Medicina. 2021;57(9):868. doi:10.3390/medicina57090868
3. Su S, Wang W, Sun T, et al. Smoking as a risk factor for diabetic nephropathy: a meta-analysis. Int Urol Nephrol. 2017;49(10):1801–1807. doi:10.1007/s11255-017-1638-3
4. Samsu N. Diabetic nephropathy: challenges in pathogenesis, diagnosis, and Treatment. Biomed Res Int. 2021;2021:1497449. doi:10.1155/2021/1497449
5. Saeed M. Locus and gene-based GWAS meta-analysis identifies new diabetic nephropathy genes. Immunogenetics. 2018;70(6):347–353. doi:10.1007/s00251-017-1044-0
6. Festa A, D’Agostino R, Howard G, Mykkänen L, Tracy RP, Haffner SM. Inflammation and microalbuminuria in nondiabetic and type 2 diabetic subjects: the insulin resistance atherosclerosis study. Kidney Int. 2000;58(4):1703–1710. doi:10.1046/j.1523-1755.2000.00331.x
7. Navarro JF, Mora C, Maca M, Garca J. Inflammatory parameters are independently associated with urinary albumin in type 2 diabetes mellitus. Am J Kidney Dis. 2003;42(1):53–61. doi:10.1016/S0272-6386(03)00408-6
8. Ferenbach D, Kluth DC, Hughes J. Inflammatory cells in renal injury and repair. Semin Nephrol. 2007;27(3):250–259. doi:10.1016/j.semnephrol.2007.02.001
9. Lim AK, Ma FY, Nikolic-Paterson DJ, Kitching AR, Thomas MC, Tesch GH. Lymphocytes promote albuminuria, but not renal dysfunction or histological damage in a mouse model of diabetic renal injury. Diabetologia. 2010;53(8):1772–1782. doi:10.1007/s00125-010-1757-1
10. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. doi:10.1093/hmg/ddu328
11. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333–2355. doi:10.1177/0962280215597579
12. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408.
13. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740–752. doi:10.1093/ije/dyq151
14. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi:10.1038/s41588-018-0099-7
15. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi:10.1002/gepi.21758
16. Burgess S, Thompson SG. Improving bias and coverage in instrumental variable analysis with weak instruments for continuous and binary outcomes. Stat Med. 2012;31(15):1582–1600. doi:10.1002/sim.4498
17. Yaribeygi H, Atkin SL, Sahebkar A. Interleukin-18 and diabetic nephropathy: a review. J Cell Physiol. 2019;234(5):5674–5682. doi:10.1002/jcp.27427
18. Nosratabadi R, Arababadi MK, Hassanshahi G, et al. Evaluation of IFN-gamma serum level in nephropatic type 2 diabetic patients. Pak J Biol Sci. 2009;12(9):746–749. doi:10.3923/pjbs.2009.746.749
19. Fathy SA, Mohamed MR, Ali MAM, El-Helaly AE, Alattar AT. Influence of IL-6, IL-10, IFN-γ and TNF-α genetic variants on susceptibility to diabetic kidney disease in type 2 diabetes mellitus patients. Biomarkers. 2019;24(1):43–55. doi:10.1080/1354750X.2018.1501761
20. Moon JY, Jeong KH, Lee TW, Ihm CG, Lim SJ, Lee SH. Aberrant recruitment and activation of T cells in diabetic nephropathy. Am J Nephrol. 2012;35(2):164–174. doi:10.1159/000334928
21. Kim SR, Im JE, Jeong JH, et al. The cKit inhibitor, masitinib, prevents diabetes-induced retinal vascular leakage. Invest Ophthalmol Vis Sci. 2016;57(3):1201–1206. doi:10.1167/iovs.15-18065
22. El-Asrar AM A, Struyf S, Opdenakker G, Van Damme J, Geboes K. Expression of stem cell factor/c-kit signaling pathway components in diabetic fibrovascular epiretinal membranes. Mol Vis. 2010;16:1098–1107.
23. Kelly DJ, Zhang Y, Gow RM, Itescu S, Gilbert RE. Cells expressing the stem cell factor receptor, c-kit, contribute to neoangiogenesis in diabetes. Diab Vasc Dis Res. 2005;2(2):76–80. doi:10.3132/dvdr.2005.013
24. Zent R, Pozzi A. Angiogenesis in diabetic nephropathy. Semin Nephrol. 2007;27(2):161–171. doi:10.1016/j.semnephrol.2007.01.007
25. Yin DD, Luo JH, Zhao ZY, Liao YJ, Li Y. Tranilast prevents renal interstitial fibrosis by blocking mast cell infiltration in a rat model of diabetic kidney disease. Mol Med Rep. 2018;17(5):7356–7364. doi:10.3892/mmr.2018.8776
26. McFadden C, Morgan R, Rahangdale S, et al. Preferential migration of T regulatory cells induced by IL-16. J Immunol. 2007;179(10):6439–6445. doi:10.4049/jimmunol.179.10.6439
27. Vendrame F, Cataldo D, Ciarlo L, Umland O, Misasi R, Dotta F. In type 1 diabetes immunocompetent cells are defective in IL-16 secretion. Scand J Immunol. 2012;75(1):127–128. doi:10.1111/j.1365-3083.2011.02630.x
28. Xu J, Su HL, Wang JH, Zhang CH. CD4+CD25+Foxp3+调节性T细胞与2型糖尿病肾病的关系 [Role of CD4+CD25+Foxp3+ regulatory T cells in type 2 diabetic nephropathy]. Nan Fang Yi Ke Da Xue Xue Bao. 2009;29(1):137–139. Chinese.
29. Eller K, Kirsch A, Wolf AM, et al. Potential role of regulatory T cells in reversing obesity-linked insulin resistance and diabetic nephropathy. Diabetes. 2011;60(11):2954–2962. doi:10.2337/db11-0358
30. Bystry RS, Aluvihare V, Welch KA, Kallikourdis M, Betz AG. B cells and professional APCs recruit regulatory T cells via CCL4. Nat Immunol. 2001;2(12):1126–1132. doi:10.1038/ni735
31. Patterson SJ, Pesenacker AM, Wang AY, et al. T regulatory cell chemokine production mediates pathogenic T cell attraction and suppression. J Clin Invest. 2016;126(3):1039–1051. doi:10.1172/JCI83987
32. Purohit S, Sharma A, Hopkins D, et al. Large-scale discovery and validation studies demonstrate significant reductions in circulating levels of il8, il-1ra, mcp-1, and mip-1β in patients with type 1 diabetes. J Clin Endocrinol Metab. 2015;100(9):E1179–E1187. doi:10.1210/JC.2015-1388
33. Meagher C, Arreaza G, Peters A, et al. CCL4 protects from type 1 diabetes by altering islet beta-cell-targeted inflammatory responses. Diabetes. 2007;56(3):809–817. doi:10.2337/db06-0619
34. Piani F, Melena I, Tommerdahl KL, et al. Sex-related differences in diabetic kidney disease: a review on the mechanisms and potential therapeutic implications. J Diabetes Complications. 2021;35(4):107841. doi:10.1016/j.jdiacomp.2020.107841
35. Ahola-Olli AV, Würtz P, Havulinna AS, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100(1):40–50. doi:10.1016/j.ajhg.2016.11.007
36. Wang L, Wang HL, Liu TT, Lan HY. TGF-beta as a master regulator of diabetic nephropathy. Int J Mol Sci. 2021;22(15):7881. doi:10.3390/ijms22157881
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.