Back to Journals » OncoTargets and Therapy » Volume 13
IFI30 Is a Novel Immune-Related Target with Predicting Value of Prognosis and Treatment Response in Glioblastoma
Authors Zhu C, Chen X, Guan G, Zou C, Guo Q, Cheng P, Cheng W, Wu A
Received 3 November 2019
Accepted for publication 19 December 2019
Published 5 February 2020 Volume 2020:13 Pages 1129—1143
DOI https://doi.org/10.2147/OTT.S237162
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Chen Zhu,* Xin Chen,* Gefei Guan, Cunyi Zou, Qing Guo, Peng Cheng, Wen Cheng, Anhua Wu
Department of Neurosurgery, The First Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Anhua Wu
Department of Neurosurgery, The First Hospital of China Medical University, Nanjing Street 155, Heping District, Shenyang 110001, People’s Republic of China
Tel +86 24-83283301
Fax +86 24-83283133
Email [email protected]
Wen Cheng
Department of Neurosurgery, The First Hospital of China Medical University, Nanjing Street 155, Heping District, Shenyang 110001, People’s Republic of China
Tel +86 24-83283129
Fax + 86 24-83283133
Email [email protected]
Purpose: As a crucial part of anti-tumor immunotherapy, interferon-α/β (IFN-α/β) treatment has been broadly applied to clinical trials of glioma. However, less is known about implement of interferon-γ (IFN-γ) in glioma. Further investigating the valuable hub molecular of IFN-γ family might provide us a novel guidance for glioma therapy.
Methods: This study carried out an analysis on glioma patients from the Chinese Glioma Genome Atlas (CGGA) and The Cancer Genome Atlas (TCGA) cohorts. The analyses were performed by GraphPad Prism 8 and R language. All the validated experiments were performed three times independently.
Results: We identified IFI30 as the most stable independent prognostic gene among 20 classical IFN-γ stimulated genes (ISGs) in glioma patients. Furthermore, we found that IFI30 highly expressed in malignant subtypes of glioma and associated with chemotherapy response. We also found IFI30 could activate IL6-STAT6 signal pathway to decline the glioma cells’ chemotherapy sensitivity by performing experiments. Gene ontology (GO) analysis showed IFI30 associated with enhanced leucocyte mediated immune and inflammatory response. Microenvironment analysis referred that high IFI30 expression accompanied with more infiltration of M2 type macrophages.
Conclusion: IFI30 is involved in the malignant progression and chemotherapy response of glioblastoma, which can be a potential target for treatment in glioblastoma patients.
Keywords: glioblastoma, IFI30, prognosis, microenvironment, temozolomide
Introduction
Glioblastoma (GBM) is the most common malignant tumor of the central nervous system (CNS) with the characteristics of high incidence rates and poor prognosis.1 Although the clinical approaches to the diagnosis and treatment of GBM improving constantly, there still occupies an unsatisfying median survival of 15 months2 under the standard treatment including surgery, temozolomide (TMZ) chemotherapy and radiation.3 However, in the wake of the discovery of the lymphatic system in the CNS, a novel treatment strategy, immunotherapy, brought hope for glioma patients. Increasingly, immunological studies of glioma have been carried on in order to explore effective therapies.
GBM possesses a complex tumor microenvironment consisting of endothelial cells, pericytes, fibroblasts and some immune cells. In glioma microenvironment, immune cells in particular serve as a significant regulator in tumor malignancies.4 Interferons (IFNs) are universally recognized as one of the foremost families in regulating immune response. To date, type I interferon, interferon-α/β (IFN-α/β) has been extensively used either alone or accompany with other agents in malignant glioma treatment and was reported to re-sensitize resistant glioma cells to TMZ.5–8 While another study indicated that the type II interferon, interferon-γ (IFN-γ) score was positively correlated with PDL1 expression resulting in an immune checkpoint blockage in GBM.9 The upregulation of PDL1 might further exert an influence on glioma chemotherapy sensitivity.10 Taking these into consideration, we assumed that IFN-γ might play a distinctive role in GBM progression compared to IFN-α/β. Hence, it is vital to investigate the clinical part of IFN-γ in GBM, hoping to improve the efficacy of GBM therapy.
In this study, we summarized 20 classical IFN-γ stimulated genes (ISGs) and identified IFI30 as a significantly prognostic gene in glioma among ISGs. Further studies defined IFI30’s relationship with tumor malignancy and enhanced immune response and the effects and molecular mechanism of knocking down of IFI30 on re-sensitize to TMZ were finally explored, which offered novel insights for GBM’s chemotherapy.
Materials and Methods
Ethics
This study was approved by the Ethics Committee of the First Hospital of China Medical University. 12 clinical samples information was collected from the First Hospital of China Medical University. Informed consent was obtained from each patient and was written informed consent, which was conducted in accordance with the declaration of Helsinki.
Data Collection
We collected 425 GBM patients and 875 lower grade glioma (LGG) patients for further study. Clinical traits information along with transcriptome sequencing data of CGGA microarray and RNA-seq cohorts were obtained from the Chinese Glioma Genome Atlas (CGGA) dataset (http://www.cgga.org.cn). 669 glioma patients’ clinical information and transcriptome sequencing data of The Cancer Genome Atlas (TCGA) were downloaded from https://portal.gdc.cancer.gov/. A series of mentioned clinical traits were illustrated in Supplementary Table S4. The GBM mutation data (MAF file) were downloaded from https://portal.gdc.cancer.gov/ and somatic copy number variation (SCNA) information was got from Firehose (http://gdac.broadinstitute.org/). Overall survival (OS) was estimated from diagnosis date to final follow-up or death. Methods for sequencing, MGMT promoter methylation and isocitrate dehydrogenase (IDH) mutation state were mentioned earlier.11,12 The normalization was conducted by log2 transformed after adding a 0.5 pseudocount based on the pre-processed data.
Bioinformatics Analysis
Heatmap and principal component analysis (PCA) were made by R language to distinguish different group information. Maftools R packages were performed to illustrate significant mutation information. Limma R package was used to calculate the differential CNV genes between high and low IFI30 groups which were demonstrated by Rcircos packages. The gene ontology (GO) enrichment analysis was carried out by ClueGo to find the IFI30 functional implications. Gene set enrichment analysis (GSEA) was conducted to explore differential signal pathway, immune response and regulation of leukocyte mediated immunity between patients with low and high IFI30. ESTIMATE R package was performed to calculate stromal and immune scores and GBM purity was calculated according to the formula from Yoshihara et al.13 The relative immune cells proportions were calculated based on the CIBERSORT algorithm.14 Corrplot R package was used to analyze IFI30 relationship with immune checkpoint and anti-inflammatory genes. The information of IFI30’s correlation with M2 marker CD163 in different tumors was downloaded from Timer (https://cistrome.shinyapps.io/timer/).
Cell Cultures and Materials
Human brain cancer cell lines U87MG, 229MG, 251MG, T98MG and Normal Human Astrocytes (NHA) were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) combined with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. IFI30 knockdown small-interfering RNA (siRNA) and the negative control RNA (siNC) were obtained from Sangon Biotech (Shanghai, China). Lipofectamine 3000 (Life Technologies, USA) was used for siRNAs transfection according to the manufacturer’s instructions. IFN-γ was bought from Proteintech.
Migration and Invasion Assays
The transwell chambers coated with and without 80 µL diluted Matrigel matrix were used to detect cell invasion or migration capacity. A volume of 200 µL 0.2% FBS consisting of 105 or 5×104 cells/mL after transfection was added to the upper chamber, while 600 µL media with 20% FBS was put in the bottom chamber in triplicate per group. After incubation for 22 h, 4% formaldehyde was used to fix the cells followed with 0.1% crystal violet staining for 20 mins. Then upright Microscope was used for cells observation and photography.
Real-Time Quantitative PCR (qPCR)
Total RNA from U87 cells was extracted by TaKaRa MiniBEST Universal RNA Extraction Kit. NanoDrop 2000 was used to qualify the RNA concentration. Extracted RNA was then reverse transcribed to cDNA in a 20-µL reaction system. The real-time quantitative PCR reactions were performed as follows: denaturation at 95°C for 3 min and 50 cycles of 95°C for 5 s and 60°C for 30s. Each sample was devided in triplicate. PCR primer sequences showed as follows:
IFI30 (forward primer: 5ʹ-GACCGAGAAACTGAGCTCCCC-3ʹ, reverse primer: 5ʹ-TGGCATCGAACATCTGCTGG-3ʹ); glyceraldehyde 3-phosphate dehydrogenase (GAPDH; forward primer: 5ʹ-TGACTTCAACAGCGACACCCA-3ʹ, reverse primer: 5ʹ-CACCCTGTTGCTGTAGCCAAA-3ʹ). GAPDH was identified as an internal control to calculate the relative expression of other mRNA via the 2−ΔΔCT method.
Western Blot
Transfected U87 cells were lysed by a mixture of RIPA buffer and PMSF at the concentration of 10:1. Equal amounts (20 µg) of different proteins were put in 10% gel for electrophoresis and then transferred to polyvinylidene fluoride membranes. After being blocked with 5% milk/Tris-buffered saline plus Tween 20, membrane was incubated with primary antibodies against IFI30 (1:1000; no.ab232915, Abcam), STAT6 (1:1000; no.ab32520, Abcam), P-STAT6 (1:1000; no. 9361T, Cell Signaling Technology), IL-6 (1:1000; no. #6708, R&D) and GAPDH (1:1000; no.10494-1-AP, Proteintech) at 4°C overnight. The second day, TBST was added to wash primary antibodies followed with HRP goat anti-mouse IgG and HRP goat anti-rabbit IgG (Proteintech; 1:5000) as secondary antibody incubation 1h. Immunoreactive bands were visualized by Tanon 5200. GAPDH antibody was defined as internal control.
Immunohistochemical (IHC)
The expression of IFI30 tissue proteins was detected by immunohistochemistry (IHC). We collected 12 paraffin-embedded tissues from our patients. After a graded series of alcohol dewaxing and rehydration, antigen retrieval was carried out in citrate buffer at PH=6 for 2min. Endogenous peroxidase was blocked by incubation with 3% H2O2 for 12min at room temperature. Primary antibodies against IFI30 (1:100; Abcam, ab232915) was added at 4°C followed by overnight incubation and then incubated with secondary antibody. The results were independently analyzed by two investigators based on the German Immunohistochemical Score (GIS).15
Immunofluorescence
The expression of IFI30 and CD163 (1:100, Proteintech, 16646-1-AP) was detected by immunofluorescence assay. Frozen human tissue sections were washed three times in PBS. Then, 0.1% Triton X-100 was used to permeabilize the sections for 10 min and goat serum were performed to block at 37°C for 1 h. Blocked tissues were incubated in IFI30 and CD163 antibodies at 4°C overnight. On the second day, 0.1% PBS-Tween 20 (PBS-T) was used to wash and then stained with suitable TRITC red-conjugated or FITC green-conjugated secondary antibodies. Finally, the sections were incubated for 10 mins in 4ʹ6-diamidino-2-phenylindole (DAPI, 10ug/mL, Sigma-Aldrich) to stain the cell nuclei and photo imaged by fluorescence microscope. The images were merged digitally to monitor the co-localization condition.
Statistical Analysis
R 3.5.2 (https://www.r-project.org/) and GraphPad Prism 8 were mainly used for statistical analysis. Kaplan-Meier survival analysis was used to evaluate the prognostic value. Univariate and multivariate Cox regression analyses were used to verify independent prognostic factors. The receiver operating characteristic (ROC) curve was made using Medcalc software. Student’s t test was performed to analyse the differential expression. Survival curve and forest plot were exported by GraphPad Prism 7. Two-tailed p value<0.05 was termed as significant.
Results
Expression of ISGs Was Dysregulated in Glioma
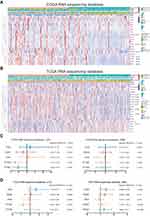
As the critical role of IFN-γ in immune regulation and tumor development, we summarized 20 classical IFN-γ stimulated genes and explored their prognostic value in glioma (Supplementary Table S1). The glioma patients of CGGA RNA-seq database were stratified into two groups according to their survival days. We ranked these patients by survival days and depicted these 20 ISGs expression pattern by heatmap (Figure 1A). We compared their expression between long and short survival groups, the results showed seven ISGs were up-regulated significantly in short survival group, included IFITM1, IFI44, ISG20L2, ITITM2, IFITM3, ISG20 and IFI30. Then, we conducted a similar analysis in the TCGA RNA-seq database and found nine genes with a distinct expression between long and short survival groups (Figure 1B). There were five intersecting genes between these two cohorts (ISG20, IFI30, IFI44, IFITM2, IFITM3), which indicated that they may be associated with the poor prognosis of glioma.
IFI30 Was Identified as the Most Significant ISG Gene with Prognostic Value in GBM
To dig out an ISG with the most stable prognostic value, we performed log-rank survival analysis in LGG and GBM of CGGA RNA-seq and TCGA RNA-seq databases, respectively. The results showed only IFI30 with a stable prognostic value in both LGG and GBM of these two cohorts (Figure 1C and D). To further assess IFI30 prognostic value, patients were stratified into two groups based on IFI30 median expression value. Patients with higher IFI30 generally had shorter OS than those with lower IFI30 levels in three cohorts (Supplementary Figure S1A–C). Furthermore, for both LGG and GBM, the high IFI30 group suffered a reduced survival time compared to the low IFI30 group significantly (Supplementary Figure S1D–I).
To estimate whether IFI30 could serve as an independent prognostic factor for GBM. Univariate and multivariate regression cox analysis were conducted. The results showed that IFI30 independently indicated unfavorable prognosis in glioma when adjusted for age, KPS, MGMT promoter status, radiotherapy and chemotherapy (Table 1). Meanwhile, IFI30 expression value remained independent prognosis in the other two cohorts (Supplementary Table S2 and S3). Collectively, these findings indicated IFI30 was a robust independent factor for predicting GBM survival.
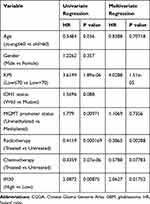 |
Table 1 Cox Regression Analysis of CGGA RNA Sequencing Database, GBM |
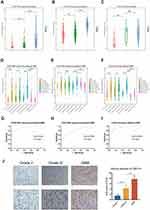
IFI30 Was Exhibited with an Expression Preference of Malignant Subtypes of GBM
To verify the association between IFI30 expression with other clinical parameters, firstly we compared IFI30 expression between Grade II, III and GBM patients. We found expression of IFI30 was positively associated with the WHO grade. (Figure 2A–C). Moreover, we found that IFI30 expression value elevated in GBM patients with IDH1 wild-type, MGMT promoter unmethylated as well as mesenchymal sub-type, recognized as more aggressive subtypes of glioma (Figure 2D–F). Further analysis of ROC curves demonstrated that IFI30 was a good predictor of mesenchymal sub-type in three cohorts (Figure 2G–I).
Then, we further explored the IFI30 expression value in glioma specimens from 12 patients using IHC, the results showed that IFI30 significantly elevated in the higher grade of gliomas (Figure 2J). These results suggested that IFI30 was expressed with a preference in aggressive subtypes of glioma, which might be involved with the malignancy of glioma.
The Prognostic Value of IFI30 Stratified by IDH1 and MGMT Promoter Status in GBM
IDH1 mutation and MGMT promoter methylation status are two main factors driving the malignancy of GBM. To explore the association between IFI30’s prognostic value and IDH1 as well as MGMT promoter status, we conducted survival analysis stratified by these two characters. Interestingly, we found there was a robust survival difference between low and high IFI30 expression groups in IDH1 wild-type patients, but not in those patients with IDH1 mutation (Figure 3A and B). Further, we investigated the relevance between IFI30 and MGMT promoter methylation. High IFI30 expression patients exhibited a significant worse survival compared with low ones just in GBM with MGMT promoter methylation, but not in those patients with MGMT promoter unmethylated (Figure 3C and D). Similar results were obtained in TCGA RNA-seq cohort and CGGA microarray cohort (Figure 3E–L).
High IFI30 Expression-Specific Somatic Mutations and Copy Number Alterations
To investigate the difference between high and low IFI30 groups at the genomic level, we analyzed somatic mutation and copy number variation (CNV) from the TCGA dataset. The common somatic mutation information of GBM stratified by IFI30 expression level was illustrated in Figure 4A. By focusing the FDR ≤ 0.05, we identified five significant somatic mutation genes. In the high IFI30 expression group, somatic mutation profiles revealed a high mutation frequency in RB1, while IDH1, ATRX, PDGFRA and MROH2B mutated more frequently in the low IFI30 group (Figure 4B). To characterize IFI30-specific SCNAs, we identified differential copy number variation genes between low and high IFI30 groups (Figure 4C). High IFI30 groups existed Chromosome 5, 22 and X partial fragments amplification and Chromosome 8 partial fragments deletion (Figure 4D). Although there were no significant differences in the copy number of some important chromosome fragments like 1p and 19q between two groups, some amplified genes in high IFI30 level like NRIP1 and SSBP2 might participate in the promotion of GBM malignant progression.16,17
High IFI30 Expression Conferred Chemotherapy Resistance in GBM
To investigate the relationship between IFI30 expression and treatment response, different treatment conditions of survival analysis were performed. In GBM patients with chemotherapy, the low IFI30 group had a survival advantage compared with the high IFI30 group (Figure 5A). However, there was no significant difference between high and low IFI30 groups in GBM patients without chemotherapy (Figure 5B). Moreover, in the low IFI30 group, patients under chemotherapy treatment lived longer than those patients without chemotherapy significantly (Figure 5C). However, this therapeutic benefit was impaired in the high IFI30 group (Figure 5D). MGMT promoter methylation is well established for implying better prognosis and chemotherapy sensitivity in GBM patients.18 We further added MGMT promoter status to survival analysis. For all GBM patients or who received chemotherapy, only MGMT promoter-methylated patients with lower IFI30 had a survival advantage over the unmethylated ones. Nevertheless, the survival time of MGMT promoter-methylated patients with high IFI30 was just similar to that of unmethylated patients (Figure 5E and F). Further, we found that even in GBM patients with MGMT promoter methylated, only patients with low IFI30 could benefit from additional chemotherapy (Figure 5G and H). Similar results were obtained in CGGA microarray cohort and TCGA RNA-seq cohort for validation (Supplementary Figure S2A-J). These results indicated that high IFI30 patients might decrease chemotherapy response even with MGMT promoter methylation.
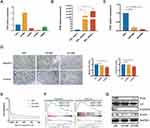
IFI30 Contributed to a Malignant Phenotype and Chemoresistance of GBM Cells
To further explore IFI30 functions involved in GBM malignant progression, IFI30 mRNA level was detected in glioma cell lines. We identified high levels of IFI30 mRNA in U87 cells (Figure 6A), which was termed as a GBM cell line with the characteristics of mesenchymal sub-type.19 We next confirmed IFN-γ truly induced IFI30 overexpression in U87 cells significantly (Figure 6B). Subsequently, siRNA was constructed and verified by qPCR (Figure 6C). To examine the malignant role of IFI30, transwell was carried out to investigate the migration and invasion of U87 cells transfected with siIFI30. Compared to the siNC group, the numbers of migration and invasive cells in the siIFI30 groups were significantly decreased (Figure 6D). In order to identify the role of IFI30 in chemotherapy, we performed various conditions U87 cells treated with different concentration of TMZ. After 72h incubation, siIFI30 U87 cells demonstrated obviously decreased viability on the TMZ concentration above 50μM compared to the siNC group (Figure 6E). To further explore the molecular mechanism mediated by IFI30, we conducted GSEA and identified that interleukin 6 (IL6) and signal transducer and activator of transcription (STAT) signal pathway enriched distinctly in high IFI30 group (Figure 6F). Finally, we performed Western blotting and confirmed that IFI30 knockdown deregulated the expression of IL6 and STAT6 level, which might further regulate GBM chemotherapy sensitivity20–22 (Figure 6G).
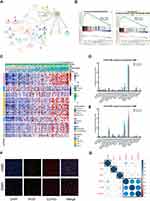
IFI30 Was Associated with Enhanced Leucocyte-Mediated Immune Response
To identify the most relevant functional biological process attributing to IFI30 high expression, we summarized one IFI30 correlated (R≥0.7) genes group. The results of GO analysis demonstrated that these genes enriched in macrophage differentiation and regulation of mononuclear cell migration (Figure 7A). GSEA further verified the biological functions of IFI30, and the results suggested that the high IFI30 group possessed enhanced immune response and regulation of leukocyte mediated immunity in the CGGA RNA-seq cohort (Figure 7B) and other two cohorts (Supplementary Figure S3A and B). Furthermore, PCA was conducted exhibiting the general distributions in different directions between low and high IFI30 groups according to “Immune Response” and “Immune Process” go terms (Supplementary Figure S3C and D), suggesting a different immune status in these two groups.
An inflammatory response is usually accompanied by immune response leading to a malignant progression of GBM. Based on the relationship between IFI30 and activated immune response, we further examined the IFI30 association with various inflammatory activity signatures.23 IFI30 was positively correlated with six of these clusters except IgG in the CGGA RNA-seq cohort (Figure 7C) and other two cohorts (Supplementary Figure S3E and F), implying an involvement in macrophage activation, T cell-related signaling transduction and antigen-presenting cells. These findings indicated that high IFI30 condition was accompanied with enhanced immune response and leukocyte related process.
The Association Between IFI30 Expression and Immune Microenvironment
To reveal the influence of IFI30 on the tumor microenvironment, we calculated the tumor purity of each GBM patient using Estimate and obtained immune microenvironment information from CIBERSORT. We found IFI30 exhibited a negative correlation with glioma purity (Supplementary Figure S3G and H). To verify the comprehensive alteration of immune microenvironment attributing to IFI30 expression, we compared the relative ratio of 22 kinds of immune cells grouped by IFI30 expression, the results showed that M0 macrophage was enriched in high IFI30 group in CGGA cohort (Figure 7D and Supplementary Figure S3I), although there was no obvious difference of M0 macrophage in TCGA sequence cohort, high IFI30 still conferred a high M2 macrophage composition (Figure 7E), which was termed as a kind of pro-tumor macrophage. Furthermore, we found that M2 macrophage marker CD163 was positively correlated with IFI30 level in pan-cancer (Supplementary Figure S4A–L). The co-location and co-expression relationships of IFI30 and CD163 were verified in GBM samples by conducting immunofluorescence (Figure 7F).
Due to the significant functions of immune checkpoint molecules in the immune process regulation, we finally assessed the relationship between IFI30 and several well-known immune checkpoint genes in GBM samples. IFI30 was highly correlated with Hepatitis A virus cellular receptor 2 (HAVCR2), also known as mucin domain-3 (TIM-3) based on the three cohorts (Figure 7G and Supplementary Figure S3J and K). To further clarify the immunosuppressive effects of IFI30 in glioma, we used correlation analysis between IFI30 and seven inhibitory molecules (TGFβ1, IL10, VEGFB, IL6, FCGR2B, MMP9, CCL18) and identified TGFβ1 and CCL18 strongly correlated with IFI30, which might further participate in M2 macrophage promoted malignant behavior (Supplementary Figure S5). Taken together, these results indicated that IFI30 acted a critical role in regulating the immune microenvironment leading to the malignant progression of glioma.
Discussion
TMZ is usually used as a frontline chemotherapy agent to treat glioma. However, considering the fact of chemoresistance of glioma cells to TMZ, the treatment is still unsatisfactory even in those patients with MGMT promoter methylated.24 Despite the accumulative researches on the relationship between genetic factors and chemoresistance,25,26 the potential molecular mechanism of chemotherapy resistance in GBM is still largely unknown. Hence, investigation on the strategies to overcome TMZ resistance and finding new targets in addition to MGMT promoter methylation is urgently needed.
Evidence has demonstrated that interferons play a significant role in immune regulation.27 Although the treatment of interferons has made great headway, especially combined with some antitumor drugs, there still existed some side effects and poor efficacy reports.28 Moreover, most interferons treatment focuses on the IFN-α/β treatment, less is known about the effect of IFN-γ applied in tumor therapy, especially in the chemotherapy. Therefore, clearly distinguishing the two types of interferons in tumor treatment and exploring the critical role of IFN-γ in chemotherapy is necessary.
Increasing researches have indicated that interferon related genes could serve as biomarkers to predict patients’ prognosis.29–31 In this study, we identified IFI30 as the most stable prognostic gene among ISGs in glioma. Importantly, we explored IFI30 crucial roles in clinical, molecular, and biological situations of GBM. IDH1 mutation is a crucial molecular event in glioma and predicts a better prognosis in GBM. While in IDH1 wild-type GBM patients, more immune cells infiltrate than the IDH1-mutant group. We found IFI30 highly expressed in IDH1 wild-type and more IDH1 mutation enrichment in the low IFI30 group. These findings indicate that IFI30 might mediate an active immune and inflammation condition in glioma.
The tumor microenvironment, especially immune cell components, exerts a crucial effect on cancer progression. Our team previously reported that glioma purity was an independent factor determining overall survival.32 We found IFI30 was strongly associated with GBM purity. Furthermore, patients with high IFI30 levels accompanied enrichment of macrophages especially M2 cell, which is the most important immune cell in antitumor immunity. These findings imply that expression levels of IFI30 reflect the formation of the glioma microenvironment, which exerts a crucial role in influencing the immune response state of macrophages in glioma.
Despite the fact that IFI30 is high-expressed in some APCs like dendritic cells, B cells and macrophages, termed as an immune activator by promoting the antigen presentation process,33–36 few studies estimated the role of IFI30 in tumor cells. We found IFI30 could also be up-regulated in U87 glioblastoma cells under IFN-γ stimulation. Although the role of IFI30 in glioma malignant progression has been reported,37 its effect on gliomas of chemotherapy sensitivity is less known. We conducted somatic mutation profiles in GBM patients stratified by IFI30 expression, the results exhibited that IDH1 and ATRX mutated more frequently in low IFI30 group, increasing evidences indicated that IDH1 and ATRX wild type conferred a higher incidence of TMZ resistance,38,39 which may indicate that IFI30 also has the ability in TMZ sensitivity regulation. By performing analyses in CGGA and TCGA cohorts, we found that high IFI30 expression indeed conferred chemotherapy resistance in GBM. Further, we found that IFI30 could down-regulate TMZ chemotherapy sensitivity in glioblastoma cells. Moreover, IL6-STAT6 pathway could be regulated by IFI30, which were termed as classical targets affecting tumor cells’ chemotherapy resistance,20–22 the above findings might offer novel insights into GBM’s chemotherapy.
Conclusions
In summary, we identified IFI30 as the most stable prognostic gene of ISGs in glioma. Further analysis indicated IFI30 could affect malignant biological behaviors of GBM and was validated by experiments. Moreover, the IL6-STAT6 signal pathway could be regulated by IFI30, which might be the mechanism of IFI30 mediating chemotherapy sensitivity decline. Furthermore, we found there existed a close relationship between IFI30 expression and IDH1 mutant status, IFI30’s overexpression often occurs in IDH1 wild-type subgroup, and commonly accompanied with a more inhibitory immune microenvironment, which was characterized with more infiltration of M2 macrophage and highly expression of inhibitory check points as well as inflammatory genes. However, this study is limited due to the lack of comprehensive experimental data. Meanwhile, the specific molecular mechanism of IFI30-mediated GBM’s chemotherapy response should be fully estimated for its application and translation.
Ethical Approval
This study was approved by the Ethics Committee of the First Hospital of China Medical University. Twelve clinical samples information was collected from the First Hospital of China Medical University. Informed consent was obtained from each patient.
Data Sharing Statement
The datasets used and/or analyzed during the current study are obtained from the Chinese Glioma Genome Atlas (CGGA) dataset (http://www.cgga.org.cn) and The Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.cancer.gov/).
Acknowledgments
We thank all the members of Dr Wu AH’s lab for helping in our study.
Author Contributions
Conceptualization, Chen Zhu, Xin Chen and Wen Cheng; Data curation, Gefei Guan, Cunyi Zou, Qing Guo; Funding acquisition, Anhua Wu, Peng Cheng, Wen Cheng; Investigation, Chen Zhu and Xin Chen; Methodology, Cunyi Zou, Xin Chen; Project administration, Chen Zhu, Xin Chen; Supervision, Anhua Wu, Wen Cheng; Validation, Chen Zhu; Writing – original draft, Chen Zhu and Xin Chen; Writing – review & editing, Anhua Wu. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Louveau A, Smirnov I, Keyes TJ, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337–341. doi:10.1038/nature14432
2. Johnson DR, HE L, JH U. Glioblastoma survival in the United States improved after Food and Drug Administration approval of bevacizumab: a population-based analysis. Cancer. 2013;119(19):3489–3495. doi:10.1002/cncr.28259
3. Quail DF, Bowman RL, Akkari L, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science (New York, NY). 2016;352(6288):aad3018. doi:10.1126/science.aad3018
4. Gieryng A, Pszczolkowska D, Walentynowicz KA, WD R, Kaminska B. Immune microenvironment of gliomas. Lab Invest. 2017;97(5):498–518. doi:10.1038/labinvest.2017.19
5. Jonasch E, Haluska FG. Interferon in oncological practice: review of interferon biology, clinical applications, and toxicities. Oncologist. 2001;6(1):34–55. doi:10.1634/theoncologist.6-1-34
6. Wakabayashi T, Natsume A, Hashizume Y, Fujii M, Mizuno M, Yoshida J. A Phase I clinical trial of interferon-beta gene therapy for high-grade glioma: novel findings from gene expression profiling and autopsy. J Gene Med. 2008;10(4):329–339. doi:10.1002/jgm.v10:4
7. Groves MD, Puduvalli VK, Gilbert MR, et al. Two Phase II trials of temozolomide with interferon-alpha2b (pegylated and non-pegylated) in patients with recurrent glioblastoma multiforme. Br J Cancer. 2009;101(4):615–620. doi:10.1038/sj.bjc.6605189
8. Ohno M, Natsume A, Fujii M, Ito M, Wakabayashi T. Interferon-beta, MCNU, and conventional radiotherapy for pediatric patients with brainstem glioma. Pediatr Blood Cancer. 2009;53(1):37–41. doi:10.1002/pbc.v53:1
9. Qian J, Wang C, Wang B, et al. The IFN-gamma/PD-L1 axis between T cells and tumor microenvironment: hints for glioma anti-PD-1/PD-L1 therapy. J Neuroinflam. 2018;15(1):290.
10. Benci JL, Xu B, Qiu Y, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–1554.e1512. doi:10.1016/j.cell.2016.11.022
11. Cheng W, Li M, Jiang Y, et al. Association between small heat shock protein B11 and the prognostic value of MGMT promoter methylation in patients with high-grade glioma. J Neurosurg. 2016;125(1):7–16. doi:10.3171/2015.5.JNS142437
12. Cheng W, Li M, Cai J, et al. HDAC4, a prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylation. J Neurooncol. 2015;122(2):303–312. doi:10.1007/s11060-014-1709-6
13. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003
14. Newman AM, Liu CL, Green MR. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
15. Remmele W, Schicketanz KH. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Computer-assisted image analysis (QIC score) vs. subjective grading (IRS). Pathol Res Pract. 1993;189(8):862–866. doi:10.1016/S0344-0338(11)81095-2
16. Xiao Y, Decker PA, Rice T, et al. SSBP2 variants are associated with survival in glioblastoma patients. Clin Cancer Res. 2012;18(11):3154–3162. doi:10.1158/1078-0432.CCR-11-2778
17. Minchenko DO, Riabovol OO, Ratushna OO, Minchenko OH. Hypoxic regulation of the expression of genes encoded estrogen related proteins in U87 glioma cells: eff ect of IRE1 inhibition. Endocr Regul. 2017;51(1):8–19. doi:10.1515/enr-2017-0002
18. Melguizo C, Prados J, Gonzalez B, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med. 2012;10:250. doi:10.1186/1479-5876-10-250
19. Cheng W, Zhang C, Ren X, et al. Bioinformatic analyses reveal a distinct Notch activation induced by STAT3 phosphorylation in the mesenchymal subtype of glioblastoma. J Neurosurg. 2017;126(1):249–259. doi:10.3171/2015.11.JNS15432
20. Yin Y, Yao S, Hu Y, et al. The immune-microenvironment confers chemoresistance of colorectal cancer through macrophage-derived IL6. Clin Cancer Res. 2017;23(23):7375–7387. doi:10.1158/1078-0432.CCR-17-1283
21. Saha S, Mukherjee S, Khan P, et al. Aspirin suppresses the acquisition of chemoresistance in breast cancer by disrupting an NFkappaB-IL6 signaling axis responsible for the generation of cancer stem cells. Cancer Res. 2016;76(7):2000–2012. doi:10.1158/0008-5472.CAN-15-1360
22. Zhang Y, Yuan J, Zhang HY, et al. Natural resistance to apoptosis correlates with resistance to chemotherapy in colorectal cancer cells. Clin Exp Med. 2012;12(2):97–103. doi:10.1007/s10238-011-0146-5
23. Rody A, Holtrich U, Pusztai L, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi:10.1186/bcr2234
24. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised Phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi:10.1016/S1470-2045(09)70025-7
25. Reifenberger G, Wirsching HG, Knobbe-Thomsen CB and Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434–452. doi:10.1038/nrclinonc.2016.204
26. Melin B. Genetic causes of glioma: new leads in the labyrinth. Curr Opin Oncol. 2011;23(6):643–647. doi:10.1097/CCO.0b013e32834a6f61
27. Feinerman B. Tumor immunology and interferon. South Med J. 1978;71(11):1409–1411. doi:10.1097/00007611-197811000-00027
28. Yang X, Wu Y, Wang S, Huang Z, Ou W, Yu H. A randomized clinical trial on adjuvant interferon-alpha for completely resected stage I-II non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2003;6(5):339–343. doi:10.3779/j.issn.1009-3419.2003.05.04
29. Zitzmann K, Brand S, De Toni EN, et al. SOCS1 silencing enhances antitumor activity of type I IFNs by regulating apoptosis in neuroendocrine tumor cells. Cancer Res. 2007;67(10):5025–5032. doi:10.1158/0008-5472.CAN-06-2575
30. Ludigs K, Jandus C, Utzschneider DT, et al. NLRC5 shields T lymphocytes from NK-cell-mediated elimination under inflammatory conditions. Nat Commun. 2016;7:10554.
31. Lesinski GB, Zimmerer JM, Kreiner M, et al. Modulation of SOCS protein expression influences the interferon responsiveness of human melanoma cells. BMC Cancer. 2010;10:142. doi:10.1186/1471-2407-10-142
32. Zhang C, Cheng W, Ren X, et al. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23(20):6279–6291. doi:10.1158/1078-0432.CCR-16-2598
33. Hastings KT. GILT: shaping the MHC Class II-restricted peptidome and CD4(+) T cell-mediated immunity. Front Immunol. 2013;4:429. doi:10.3389/fimmu.2013.00429
34. Haque MA, Li P, Jackson SK, et al. Absence of gamma-interferon-inducible lysosomal thiol reductase in melanomas disrupts T cell recognition of select immunodominant epitopes. J Exp Med. 2002;195(10):1267–1277. doi:10.1084/jem.20011853
35. Becker JC, Schrama D. Control of central and peripheral tolerance to melanocyte differentiation antigens by GILT. J Invest Dermatol. 2012;132(1):15–17. doi:10.1038/jid.2011.361
36. Rausch MP, Hastings KT. GILT modulates CD4+ T-cell tolerance to the melanocyte differentiation antigen tyrosinase-related protein 1. J Invest Dermatol. 2012;132(1):154–162. doi:10.1038/jid.2011.236
37. Chen S, Wang Q, Shao X, et al. Lentivirus mediated gamma-interferon-inducible lysosomal thiol reductase (GILT) knockdown suppresses human glioma U373MG cell proliferation. Biochem Biophys Res Commun. 2019;509(1):182–187. doi:10.1016/j.bbrc.2018.12.099
38. Wang JB, Dong DF, Wang MD, Gao K. IDH1 overexpression induced chemotherapy resistance and IDH1 mutation enhanced chemotherapy sensitivity in Glioma cells in vitro and in vivo. Asian Pac J f Cancer Prev. 2014;15(1):427–432. doi:10.7314/APJCP.2014.15.1.427
39. Han B, Cai J, Gao W, et al. Loss of ATRX suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance temozolomide sensitivity in glioma. Cancer Lett. 2018;419:280–290. doi:10.1016/j.canlet.2018.01.056
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.