Back to Journals » Drug Design, Development and Therapy » Volume 16
Identifying the Antitumor Effects of Curcumin on Lung Adenocarcinoma Using Comprehensive Bioinformatics Analysis
Authors Yang FR, Li SY, Hu XW, Li XR , Li HJ
Received 20 May 2022
Accepted for publication 16 July 2022
Published 23 July 2022 Volume 2022:16 Pages 2365—2382
DOI https://doi.org/10.2147/DDDT.S371420
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Anastasios Lymperopoulos
Fei-Ran Yang,1,* Si-Yi Li,1,* Xi-Wen Hu,1 Xiu-Rong Li,2 Hui-Jie Li2
1College of First Clinical Medicine, Shandong University of Traditional Chinese Medicine, Jinan, Shandong, People’s Republic of China; 2Department of Oncology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, Shandong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hui-Jie Li; Xiu-Rong Li, Department of Oncology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jingshi Road, Jinan, Shandong, 250014, People’s Republic of China, Email [email protected]; [email protected]
Background: As the main component of turmeric (Curcuma longa L.), curcumin is widely used in the treatment of various diseases. Previous studies have demonstrated that curcumin has great potential as a therapeutic agent, but the lack of understanding of the functional mechanism of the drug has hindered the widespread use of the natural product. In the present study, we used comprehensive bioinformatics analysis and in vitro experiments to explore the anti-tumor mechanism of curcumin.
Materials and Methods: LUAD mRNA expression data were obtained from TCGA database and differentially expressed genes (DEGs) were identified using R software. Functional enrichment analysis was conducted to further clarify its biological properties and hub genes were identified by a protein–protein interaction (PPI) network analysis. Survival analysis and molecular docking were used to analyze the effectiveness of the hub genes. By an in vitro study, we evaluated whether curcumin could influence the proliferation, migration, and invasion activities of LUAD cells.
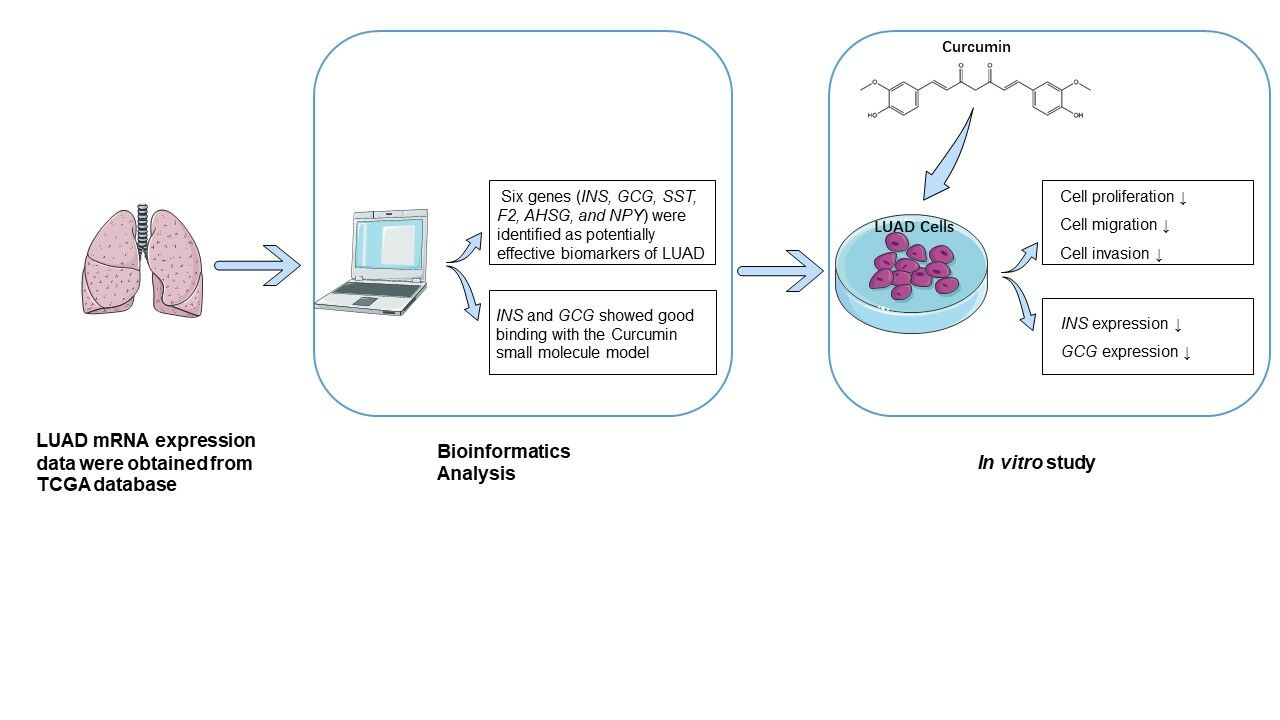
Results: In this study, 1783 DEGs from LUAD tissue samples compared to normal samples were evaluated. Functional enrichment analysis and the PPI network revealed the characteristics of the DEGs. We performed a topological analysis and identified 10 hub genes. Of these, six genes (INS, GCG, SST, F2, AHSG, and NPY) were identified as potentially effective biomarkers of LUAD. The molecular docking results indicated that curcumin targets in regulating lung cancer may be INS and GCG. We found that curcumin significantly inhibited the proliferation, migration, and invasion of LUAD cells and significantly decreased the expression of the INS and GCG genes.
Conclusion: The results of this study suggest that the therapeutic effects of curcumin on LUAD may be achieved through the intervention of INS and GCG, which may act as potential biomarkers for LUAD prevention and treatment.
Keywords: lung adenocarcinoma, curcumin, antitumor effect, bioinformatics analysis, TCGA
Graphical Abstract:
Background
Lung cancer has one of the highest incidence and mortality rates, and more than 85% are non-small cell lung cancer (NSCLC).1 Surgical resection, chemotherapy, and radiotherapy are currently the main clinical treatments for NSCLC, and for early-stage NSCLC, timely surgery can achieve a curative effect.2,3 Unfortunately, more than 70% of cancer patients present with local invasion, lymph node involvement, and distant metastases at the time of first diagnosis and are therefore ineligible for surgery.4 In recent years, many advances in the treatment of NSCLC have been made, such as tyrosine kinase inhibitors (TKIs) which are active against oncogenic alterations or immunotherapies.5 However, the development of clinically effective therapies has achieved limited success.6 Although personalized targeted therapies have been widely used in the clinic, only patients whose tumors harbor molecular mutations can benefit from this approach.7 Furthermore, despite the increasing importance of immunotherapy in the treatment of NSCLC in recent years, response rates lower than 40% have been achieved in some populations.8 Therefore, the search for new and effective therapeutic drug targets is crucial in the treatment of NSCLC.
Natural compounds derived from plants serve as a drug source and have led to the introduction of several anti-cancer drugs. For example, natural compounds derived from Taxols and vinca alkaloids have been widely used in clinical treatment, and the use of natural compounds with significant anticancer activity in combination with first-line chemotherapeutic agents has also emerged as a promising approach in cancer treatment.9 These natural compounds, alone or in combination with chemotherapeutic agents, reduce tumor growth by targeting key mediators and regulators to stop the cell cycle at different stages, by blocking anti-apoptotic proteins to induce apoptosis, or by modulating signaling pathways to induce pro-apoptotic proteins.10 Over the past decade, immunotherapy has made significant advances in the treatment of malignant tumors. Accumulated studies have revealed that natural compounds and their derivatives exert antitumor effects through their modulatory effects on the immune system, including T cells, B cells, NKs, myeloid-derived suppressor cells, immunogenic cell death, and immune checkpoints,11 which support the great potential of natural compounds for tumor therapy.
Curcuma longa L., also known as turmeric, has been widely used as a traditional medicine in China, India, and Southeast Asia. As the main component of turmeric, curcumin was first used to treat cholecystitis in 1937.12 Curcumin mediates its anticancer activity by modulating molecular targets including transcription factors, micro-RNAs, cytokines, and interfering with genes related to apoptosis and proliferation—thus inhibiting tumor cell proliferation and migration, inhibiting angiogenesis, inducing apoptosis, and increasing sensitivity to antitumor therapy.13,14 Due to its excellent pharmacological activity, curcumin is widely used in the treatment of many diseases, including gastrointestinal disease,15 liver cirrhosis,16 cardiovascular disease,17 diabetes,18 and cancer.19 Increasing studies suggest that curcumin inhibits the growth of lung cancer cells through multiple pathways by inducing apoptosis, inhibiting cell proliferation, and epigenetic changes.20 These studies have indicated that curcumin has great potential in the treatment of various diseases, but the lack of understanding of the functional mechanism of the drug hinders the wide application of natural products.
Recently, due to the rapid advances in high‐throughput technologies, especially RNA sequencing, hundreds of thousands of gene transcripts have been identified.21 Using novel computer technologies, a significant amount of data has been generated, which are publicly accessible in multiple databases. The Cancer Genome Atlas (TCGA) is the largest database to date containing tumor molecular maps of 11,160 patients from 33 cancer types, and the accumulated raw data are open for access.22 In the present study, we merged the raw data downloaded from TCGA database into a gene expression matrix and identified differentially expressed genes (DEGs). Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed and a protein–protein interaction (PPI) network of DEGs was constructed to identify 10 hub genes. We then used survival analysis and molecular docking methods to select potential targets of curcumin intervention in LUAD. Finally, we verified the validity of the bioinformatics results through in vitro experiments.
Materials and Methods
Data Source and Identification of DEGs
mRNA expression data for LUAD were downloaded from TCGA (https://cancergenome.nih.gov/),23 and consisted of 59 normal tissue samples and 535 LUAD samples. In total, 594 samples were obtained in this study. We use Perl language and Ensembl database to merge the RNA-seq raw files into a gene symbol matrix file. Subsequently, the “edgeR” package of R software was used to screen for DEGs between LUAD tissue samples and normal tissue samples.
Functional Enrichment Analysis
Functional enrichment analysis of DEGs was performed using GO and KEGG from the DAVID online database (version 6.8; https://david.ncifcrf.gov/),24 and P < 0.05 was used as a cut-off value. Finally, the “ggplot2” package was used in R software for data visualization.
PPI Network Construction
The STRING database (version 11.0; https://string-db.org/)25 was used to retrieve the predicted PPIs of the LUAD DEGs, which were visualized using Cytoscape software. The plug-in MCODE of Cytoscape was utilized to perform a module analysis of the PPI network,26 and the plug-in CytoNCA was used to calculate the degree of connectivity; the top 10 hub genes in the network were identified.27
Survival Analysis
The Kaplan–Meier Plotter database (http://www.kmplot.com) was used to perform a survival analysis of DEGs in patients with NSCLC, of which 719 patients had LUAD.28 HR with a 95% confidence interval and a logarithmic rank P-value were calculated and displayed graphically. The median expression cutoff value and P <0.05 were considered significant.
Molecular Docking
Molecular docking was used to predict the interaction between receptors and ligands and was a powerful computational tool,29 while the Surflex-Dock program in Sybyl-X 2.0 (Tripos, MO, USA) was used for molecular docking.30 The X-ray crystal structure of proteins was retrieved from the protein data bank (PDB) database, and the structure of curcumin was constructed using ChemBioOffice 15.1 (Waltham, MA, USA). Before the docking simulation started, we performed a series of modifications, including removing co-crystal ligands and structural water molecules, adding polar hydrogen atoms, termini treatment, and assigned the AMBER7 FF99 field atomic charge to protein atoms. During this docking process, based on the similarity between the structure of the ligand and the co-crystallized ligand of the target compound, the ProtoMol threshold parameter was set at 0.50. The total score ≥5 was identified as key DEGs that could be docked with curcumin. The docking results were then imported into PyMOL software to generate figures.
Cell Culture
The normal human lung epithelial cell line BEAS-2B was obtained from the ATCC, and the human LUAD cell line A549 and NCI-H1299 were obtained from the Shandong Academy of Medical Sciences (Jinan, China). All cells were cultured in RPMI-1640 medium (HyClone, UT, USA) containing 10% fetal bovine serum (Gibco, CA, USA) and placed in a humidified incubator at 37°C in an atmosphere of 95% air and 5% CO2.
Cell Viability
The cell viability was measured by a MTT assay as previously described.31 In brief, cells were seeded at a density of 1×104 cells/well and cultured in 96-well plates for 24h. The cells were then treated with different concentrations (40, 20, 10, 5, and 2.5 μM) of curcumin (purity>95%, Solarbio, Beijing, China) for 72 h at 37°C. The results of the MTT assay were used to determine the intervention concentration of curcumin in the subsequent experiments.
Wound Healing Assay
LUAD cells were seeded at a density of 1×105 cells/well in six-well plates and incubated for 24 h. Linear wounds were scraped on the cell monolayer with a 200 μL pipette tip and the cell layer was washed three times with PBS. Cells were incubated with 0, 10, and 20 μM curcumin in serum-free medium for 24 h, wounds were measured and photographed using a microscope (Carl Zeiss, Oberkochen, Germany).
Transwell Invasion Assay
The Transwell membranes were coated with Matrigel (BD Biosciences, NJ, USA) and serum-starved cells were transferred to the upper chamber of 24-well Transwell plates (8-µm pores) at a density of 1×104 cells/well, and 200 μL of serum-free RPMI-1640 medium containing curcumin (0, 10 and 20 μM). RPMI-1640 medium with 20% FBS was then added to the lower chambers as the chemoattractant. After incubation for 24 h, the chambers were removed and washed three times, and the cells were then fixed with 4% paraformaldehyde for 20 min. Finally, cells were stained with 0.1% crystal violet solution. The top layer of cells was wiped off with a wet cotton swab, and the remaining cells were counted with an optical microscope.
Quantitative Reverse Transcription PCR
qRT-PCR was performed as previously described.31 The sequences used were as follows: INS, forward: 5′- TGCATCAGAAGAGGCCATCA-3′, reverse: 5′- CGTTCCCCGCACACTAGGTA-3′; GCG, forward: 5′- TCCGATCTGACATATCTGCATT-3′, reverse: 5′- CACCACTGTGGCTACCAGTTC-3′. The Total RNA Kit I (OMEGA Biotek, GA, USA) was used to extract total RNA. PrimeScriptTM RT reagent Kit and SYBR Premix Ex Taq II kit (Takara, Shiga, Japan) were used for qRT-PCR and the Roche LightCycler® 480 was used for data analysis. Finally, the comparative CT (2−ΔΔCT) method was used to determine the relative concentration of amplified products.
Statistical Analysis
All data were analyzed using GraphPad Prism 7 (GraphPad Software, CA, USA). The results are expressed as mean ±SD, and One-way ANOVA was used for the comparison of the differences between groups; P <0.05 was considered statistically significant.
Results
Identification of DEGs in LUAD
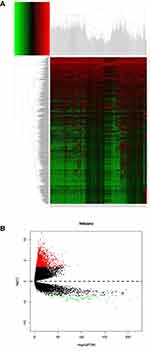
A total of 1783 DEGs, including 1704 up-regulated and 79 down-regulated genes were selected based on the criteria of | log FC |≥ 3.5 and a P-value < 0.01 (Figure 1, Table S1).
Enrichment Analysis of the DEGs
To further investigate the biological functions of the 1783 DEGs, GO analysis was applied, which included an analysis of the biological process (BP), the cellular component (CC), and the molecular function (MF) groups using the DAVID 6.8 online database and the R package “ggplot2” for data visualization. GO analysis results showed that BP is mainly enriched in the regulation of cell proliferation and transcription from the RNA polymerase II promoter, cell differentiation, cell–cell signaling and multicellular organism development; CC is associated with the extracellular region and space, an integral component of the plasma membrane, the cell junction, and the proteinaceous extracellular matrix; MF is mainly enriched in sequence-specific DNA binding, calcium ion binding, transcription factor activity, and protein homodimerization activity (Figure 2A, Table S2).
 |
Figure 2 Functional enrichment analysis of DEGs in LUAD. (A) GO analysis for DEGs. (B) KEGG pathways for DEGs. (P < 0.05). |
A total of 20 significantly enriched pathways were identified. The results demonstrated that the most enriched KEGG pathways involved neuroactive ligand-receptor interactions, the PI3K-Akt signaling pathway, steroid hormone biosynthesis, the metabolism of xenobiotics by cytochrome P450, and the Ras signaling pathway (Figure 2B, Table S3).
Identification of Hub Genes in the PPI Network
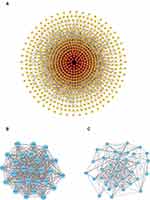
The PPI analysis of these DEGs revealed that there were 749 nodes and 3886 edges (Figure 3A). Subsequently, the two most meaningful modules were selected via the Cytoscape MCODE plug-in. Module 1 contained 27 nodes and 343 edges, enriched in the cell cycle process, ubiquitin-mediated proteolysis, and oocyte meiosis (Figure 3B). Module 2 contained 35 nodes and 209 edges and was enriched in insulin secretion, the glucagon signaling pathway and the neuroactive ligand-receptor interaction (Figure 3C) (Table S4).
The Cytoscape CytoNCA plug-in was used to perform topology analysis. Depending on the node degree, the first 10 DEGs were selected as the hub genes for subsequent analysis: albumin (ALB), insulin (INS), glucagon (GCG), coagulation factor II, thrombin (F2), ISL LIM homeobox-1 (ISL1), alpha-fetoprotein (AFP), somatostatin (SST), alpha 2-HS glycoprotein (AHSG), orthodenticle homeobox-2 (OTX2), and neuropeptide Y (NPY) (Table 1).
 |
Table 1 The Top 10 Hub Genes Identified from the Topology Analysis of the PPI Network |
Survival Analysis
The prognostic information for the 10 hub genes was explored using the online Kaplan–Meier Plotter database. The results showed that the high expression of six genes was associated with the worst OS in LUAD, including INS, GCG, SST, F2, AHSG, and NPY. While the expression of ALB, ISL1, AFP, and OTX2 were not significantly related to OS in patients with LUAD. This suggested that these six genes were potentially effective biomarkers of LUAD (Figure 4).
 |
Figure 4 Kaplan–Meier survival curves of six hub genes in patients with LUAD. Overall survival (OS) stratified by low and high expression of (A) INS, (B) GCG, (C) AHSG, (D) SST, (E) NPY, and (F) F2. |
Molecular Docking Model
Molecular docking analysis can reveal potential binding relationships between small molecules and target genes. Therefore, the six hub genes identified above were inputted into Sybyl-X 2.0 for molecular docking verification. There is a stable binding site between the protein and the small molecule model when the docking score is greater than 5,32 and the results showed that INS (PDB ID: 3ZQR, docking score: 6.8072), GCG (PDB ID: 5OTU, docking score: 6.2700) interacted well with the curcumin small molecule model, with curcumin residues having hydrogen bonding interactions at the binding site (Figure 5, Table S5).
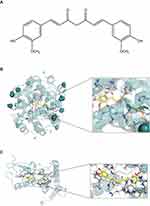 |
Figure 5 Docking combination of Curcumin and key proteins. (A) Chemical structure of curcumin. (B) In yellow, curcumin and in red, the binding site of INS and (C) GCG. |
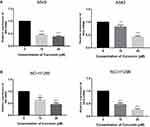
Effects of Curcumin on Cellular Viability
LUAD cells and BEAS-2B cells were treated with different concentrations (40, 20, 10, 5, and 2.5 μM) of curcumin for 24 h. The results showed that A549 and NCI-H1299 cells were more sensitive to curcumin than BEAS-2B cells (Figure 6A). Subsequently, A549 and NCI-H1299 cells were treated with curcumin for 48 and 72 h to observe the effect of curcumin on cell viability. The results showed that cell viability was related to the time of intervention as well as the concentration of curcumin (Figure 6B). The IC50 values of curcumin in A549 and NCI-H1299 cells were 15.07 and 16.71 μM at 24 h, respectively. Therefore, 10 and 20 μM were chosen as the intervention concentrations for curcumin in subsequent experiments (Figure 6C).
Cell Migration and Invasion Assays
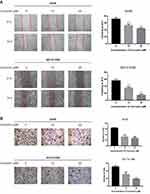
We evaluated the role of curcumin in lung cancer metastasis by cell migration and invasion assays. The results of the wound healing assay demonstrated that the curcumin 10 and 20 μM treatment groups significantly inhibited the migratory activity of A549 and NCI-H1299 cells compared with the blank control group (Figure 7A). The Transwell assay consistently showed comparable results: A549 and NCI-H1299 cells were significantly inhibited by curcumin at 10 and 20 μM (Figure 7B). The results of the wound healing and Transwell assays indicated that curcumin inhibited the migration and invasion of human LUAD cells.
Expression of INS and GCG in LUAD Cells
The results of qRT-PCR assay demonstrated that there were significant differences in INS and GCG gene expression in LUAD cells in each concentration gradient group compared with the blank control group. The results indicated that curcumin induced concentration-dependent downregulation of INS and GCG expression (Figure 8).
Discussion
NSCLC is a malignant tumor with a poor prognosis.33 Although platinum-containing combination chemotherapy is currently the main treatment for NSCLC, side effects and resistance to drugs are frequent, making chemotherapy ineffective.34 Therefore, identifying genes that are dysregulated in the development of NSCLC and further research on potentially effective drugs will help understand the molecular mechanism of NSCLC and contribute to the development of new NSCLC therapeutic targets in the future. Recently, bioinformatics analysis based on TCGA data has increased our understanding of tumor biology and has provided unprecedented opportunities for further study of tumor mechanisms.35 This was the first comprehensive analysis of the anti-lung cancer effects of curcumin using bioinformatics and in vitro studies based on A549, NCI-1299 and BEAS-2B cell lines. In this study, data analysis from TCGA database revealed 1783 DEGs, and through a topology attribution analysis of the PPI network, 10 hub genes were obtained for subsequent survival analysis. We found that the high expression of six genes (INS, GCG, SST, F2, AHSG, and NPY) was associated with the worst OS in LUAD, indicating that together, the genes may act as potential candidates for biomarkers of LUAD.
Curcumin is a phenolic compound extracted from traditional Chinese medicines. Previous studies have shown that curcumin has numerous therapeutic activities, including anticancer, anti-inflammatory, antioxidant, and anti-atherosclerosis properties.36–38 Increasing evidence has demonstrated that curcumin can exhibit antitumor activity with various molecular pathways in human lung cancer, such as through the JAK2/STAT3 signaling pathway,39 the PI3K/Akt signaling pathway40 and the Wnt/β-catenin pathway.41 Epidermal growth factor receptor (EGFR) belongs to the family of growth factor receptor tyrosine kinases and is closely related to cellular functions such as cell proliferation, dichotomization, and division. EGFR can affect tumorigenesis by activating the Raf/MEK/Erk, STAT, and PI3K/AKT pathways,42,43 and previous studies have shown that curcumin and its derivatives exert potent antitumor activity by interfering with EGFR tyrosine kinases.44,45 Curcumin decreased the overall level of cell cycle protein D1 in lung cancer cells by downregulating the expression of EGFR and thus increasing the expression of the potential tumor suppressor gene UBE1L. As a result, inhibition of cell growth was promoted.46 E-cadherin is an epithelial cell–cell adhesion molecule involved in the process of tumor epithelial mesenchymal transformation, and impairment and loss of its function is associated with the cellular characteristics of malignant transformation.47,48 Interestingly, the results of functional enrichment analysis showed that DEGs were enriched in the metabolism of xenobiotics by cytochrome P450. It has been shown that curcumin could bind to the active sites of steroid metabolizing P450s and the inhibition of CYP17A1 and CYP19A1 provides a template for structural modifications to produce effective and safe compounds to inhibit the growth of prostate cancer as well as breast cancer.49 However, there are still lack of studies on curcumin evaluating the inhibitory effects on cytochrome P450 and thus as an anti-lung cancer agent, which provides a direction for future research. Curcumin has been reported to inhibit the invasion and metastasis of many cancer cells, including lung cancer, by upregulating the expression of E-cadherin.50 In the present study, the results of the MTT assay demonstrated that curcumin reduced cell viability in a time- and dose-dependent manner. However, it should be noted that despite the high tolerance of LUAD cells to starvation, 72 h of serum-free culture could still influence cell viability. Furthermore, the results of the wound healing and Transwell assays indicated that curcumin inhibited the migration and invasion of human LUAD adenocarcinoma cells. Network pharmacology is considered to be a promising method for predicting potential new drug targets for specific diseases51,52 and molecular docking is an important approach for computer-aided drug design based on structural molecular biology.53,54 Herein, we established a molecular docking model of small molecule curcumin and the six DEGs; we found that curcumin showed a strong affinity for INS and GCG.
Insulin is produced by pancreatic islet β cells by the INS gene located at the chromosomal region 11p15.5. Elevated serum insulin concentration can inhibit autophagy, proteasome activity, and apoptosis, and lead to mitogenic effects.55–57 A study by Wang et al58 demonstrated that increased incidence of pancreatic cancer in patients with hyperinsulinemia and type 2 diabetes may be associated with increased proliferation and invasion of immortalized human pancreatic ductal cell lines by insulin. Sarkar et al59 proved that insulin increases the migration and aggressiveness of prostate cancer and that its activity is associated with decreased survival. Previous studies have also shown that INS overexpression regulates multiple downstream signal transduction pathways—including the PI3K/Akt and MAPK pathways, which are closely related to the occurrence of tumors.60–62 Glucagon is an important regulator of glucose metabolism, which is cleaved into glucagon-like peptides (GLP)-1, GLP-2, and other small peptides in enteroendocrine cells and brain neurons.63 In addition to promoting hepatic glucose production by combining with glucagon receptor on liver cells to increase blood glucose levels,64 glucagon increases lipolysis in adipose tissue, regulates glomerular filtration, reduces gastrointestinal motility, and induces proliferation of mesangial cells.65 In cancer-related studies, glucagon has been shown to enhance the proliferation of human colorectal cancer cells in vitro.66,67 In our study, the result of the qRT-PCR assay showed that curcumin inhibited the expression of INS and GCG genes, which is consistent with the results of the comprehensive bioinformatics analysis. Interestingly, high levels of insulin and glucagon in serum induce and aggravate insulin resistance.68,69 Several studies have shown that chronic hyperinsulinemia caused by insulin resistance may play an important regulatory role in the development of cancer.70,71 Especially in breast cancer, insulin resistance is a risk factor for both diabetic and non-diabetic women.72–74
Because the in vitro experiments in this study were a preliminary verification of the results of bioinformatics analysis, there are limitations that must be acknowledged. Because insulin and glucagon proteins are difficult to detect in tumor cells, we only performed a primary verification at the mRNA level. Furthermore, the present study only elucidated the correlation between curcumin and INS and GCG gene expression. The mechanisms and regulatory pathways of curcumin in the regulation of INS and GCG are still unclear, and it is also unclear whether curcumin has any direct regulatory effects on INS and GCG. Based on the results of bioinformatics analysis and results of in vitro experiments, we found that the antitumor effect of curcumin may be related to the reduction of insulin resistance in tumors; however, this hypothesis needs further studies for verification. Therefore, in our future research, we will design a series of experimental and clinical studies to investigate in depth the regulatory mechanisms of curcumin on INS and GCG and its relationship with glucose metabolism in LUAD.
Conclusions
Six genes (INS, GCG, SST, F2, AHSG and NPY) were identified as potentially effective biomarkers of LUAD by bioinformatics analysis, among which INS and GCG showed good binding with the curcumin small molecule model, and by in vitro experiments, we found that curcumin significantly inhibited the proliferation, migration and invasion of LUAD A549 and NCI-H1299 cells and significantly decreased the expression of INS and GCG genes.
Ethics Approval and Consent to Participate
TCGA belong to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Thus, Affiliated Hospital of Shandong University of Traditional Chinese Medicine Ethics Committee reviewing and waiving the need for ethical approval for the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was supported by National Natural Science Foundation of China (Grant no. 81503542) and Natural Science Foundation of Shandong Province (Grant no. ZR2021LZY029).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. doi:10.1038/nrc3775
3. Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26(21):3552–3559. doi:10.1200/JCO.2007.13.9030
4. DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. doi:10.3322/caac.21235
5. Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377(9):849–861. doi:10.1056/NEJMra1703413
6. Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19(11):1450–1464. doi:10.1038/nm.3391
7. Korpanty GJ, Graham DM, Vincent MD, Leighl NB. Biomarkers that currently affect clinical practice in lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol. 2014;4:204. doi:10.3389/fonc.2014.00204
8. Prelaj A, Tay R, Ferrara R, Chaput N, Besse B, Califano R. Predictive biomarkers of response for immune checkpoint inhibitors in non-small-cell lung cancer. Eur J Cancer. 2019;106:144–159. doi:10.1016/j.ejca.2018.11.002
9. Wattanathamsan O, Hayakawa Y, Pongrakhananon V. Molecular mechanisms of natural compounds in cell death induction and sensitization to chemotherapeutic drugs in lung cancer. Phytother Res. 2019;33(10):2531–2547. doi:10.1002/ptr.6422
10. Lin YJ, Liang WM, Chen CJ, et al. Network analysis and mechanisms of action of Chinese herb-related natural compounds in lung cancer cells. Phytomedicine. 2019;58:152893. doi:10.1016/j.phymed.2019.152893
11. Deng LJ, Qi M, Li N, Lei YH, Zhang DM, Chen JX. Natural products and their derivatives: promising modulators of tumor immunotherapy. J Leukoc Biol. 2020;108(2):493–508. doi:10.1002/JLB.3MR0320-444R
12. Hu RW, Carey EJ, Lindor KD, Tabibian JH. Curcumin in hepatobiliary disease: pharmacotherapeutic properties and emerging potential clinical applications. Ann Hepatol. 2017;16(6):835–841. doi:10.5604/01.3001.0010.5273
13. Tunc D, Dere E, Karakas D, Cevatemre B, Yilmaz VT, Ulukaya E. Cytotoxic and apoptotic effects of the combination of palladium (II) 5,5-diethylbarbiturate complex with bis(2-pyridylmethyl) amine and curcumin on non small lung cancer cell lines. Bioorg Med Chem. 2017;25(5):1717–1723. doi:10.1016/j.bmc.2017.01.043
14. Ichiki K, Mitani N, Doki Y, Hara H, Misaki T, Saiki I. Regulation of activator protein-1 activity in the mediastinal lymph node metastasis of lung cancer. Clin Exp Metastasis. 2000;18(7):539–545. doi:10.1023/A:1011980313237
15. Patel SS, Acharya A, Ray RS, Agrawal R, Raghuwanshi R, Jain P. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit Rev Food Sci Nutr. 2020;60(6):887–939. doi:10.1080/10408398.2018.1552244
16. Nouri-Vaskeh M, Malek Mahdavi A, Afshan H, Alizadeh L, Zarei M. Effect of curcumin supplementation on disease severity in patients with liver cirrhosis: a randomized controlled trial. Phytother Res. 2020;34(6):1446–1454. doi:10.1002/ptr.6620
17. Jiang S, Han J, Li T, et al. Curcumin as a potential protective compound against cardiac diseases. Pharmacol Res. 2017;119:373–383. doi:10.1016/j.phrs.2017.03.001
18. Mantzorou M, Pavlidou E, Vasios G, Tsagalioti E, Giaginis C. Effects of curcumin consumption on human chronic diseases: a narrative review of the most recent clinical data. Phytother Res. 2018;32(6):957–975. doi:10.1002/ptr.6037
19. Panda AK, Chakraborty D, Sarkar I, Khan T, Sa G. New insights into therapeutic activity and anticancer properties of curcumin. J Exp Pharmacol. 2017;9:31–45. doi:10.2147/JEP.S70568
20. Kumar G, Mittal S, Sak K, Tuli HS. Molecular mechanisms underlying chemopreventive potential of curcumin: current challenges and future perspectives. Life Sci. 2016;148:313–328. doi:10.1016/j.lfs.2016.02.022
21. Prasad NB, Somervell H, Tufano RP, et al. Identification of genes differentially expressed in benign versus malignant thyroid tumors. Clin Cancer Res. 2008;14(11):3327–3337. doi:10.1158/1078-0432.CCR-07-4495
22. Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e411. doi:10.1016/j.cell.2018.02.052
23. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1a):A68–77. doi:10.5114/wo.2014.47136
24. Jiao X, Sherman BT, Huang da W, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–1806. doi:10.1093/bioinformatics/bts251
25. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–815. doi:10.1093/nar/gks1094
26. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi:10.1186/1471-2105-4-2
27. Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Bio Syst. 2015;127:67–72. doi:10.1016/j.biosystems.2014.11.005
28. Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. doi:10.1007/s10549-016-4013-7
29. Hsin KY, Ghosh S, Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One. 2013;8(12):e83922. doi:10.1371/journal.pone.0083922
30. Jain AN. Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des. 2007;21(5):281–306. doi:10.1007/s10822-007-9114-2
31. Yang FR, Zhao YF, Hu XW, et al. Nano-realgar suppresses lung cancer stem cell growth by repressing metabolic reprogramming. Gene. 2021;788:145666. doi:10.1016/j.gene.2021.145666
32. Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46(4):499–511. doi:10.1021/jm020406h
33. David EA, Clark JM, Cooke DT, Melnikow J, Kelly K, Canter RJ. The role of thoracic surgery in the therapeutic management of metastatic non-small cell lung cancer. J Thoracic Oncol. 2017;12(11):1636–1645. doi:10.1016/j.jtho.2017.08.008
34. Kang EJ, Min KH, Hur GY, et al. Comparison of the efficacy between pemetrexed plus platinum and non-pemetrexed plus platinum as first-line treatment in patients with wild-type epidermal growth factor receptor nonsquamous non-small cell lung cancer: a retrospective analysis. Chemotherapy. 2016;61(1):41–50. doi:10.1159/000440941
35. Koboldt D, Fulton R, McLellan M, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70.
36. Sahebkar A, Serban M-C, Ursoniu S, Banach M. Effect of curcuminoids on oxidative stress: a systematic review and meta-analysis of randomized controlled trials. J Funct Foods. 2015;18:898–909. doi:10.1016/j.jff.2015.01.005
37. Um MY, Hwang KH, Choi WH, Ahn J, Jung CH, Ha TY. Curcumin attenuates adhesion molecules and matrix metalloproteinase expression in hypercholesterolemic rabbits. Nutr Res. 2014;34(10):886–893. doi:10.1016/j.nutres.2014.09.001
38. Vallianou NG, Evangelopoulos A, Schizas N, Kazazis C. Potential anticancer properties and mechanisms of action of curcumin. Anticancer Res. 2015;35(2):645–651.
39. Zhang BY, Shi YQ, Chen X, et al. Protective effect of curcumin against formaldehyde-induced genotoxicity in A549 cell lines. J Appl Toxicol. 2013;33(12):1468–1473. doi:10.1002/jat.2814
40. Jin H, Qiao F, Wang Y, Xu Y, Shang Y. Curcumin inhibits cell proliferation and induces apoptosis of human non-small cell lung cancer cells through the upregulation of miR-192-5p and suppression of PI3K/Akt signaling pathway. Oncol Rep. 2015;34(5):2782–2789. doi:10.3892/or.2015.4258
41. Lu Y, Wei C, Xi Z. Curcumin suppresses proliferation and invasion in non-small cell lung cancer by modulation of MTA1-mediated Wnt/β-catenin pathway. In Vitro Cell Dev Biol Anim. 2014;50(9):840–850. doi:10.1007/s11626-014-9779-5
42. Ye MX, Li Y, Yin H, Zhang J. Curcumin: updated molecular mechanisms and intervention targets in human lung cancer. Int J Mol Sci. 2012;13(3):3959–3978. doi:10.3390/ijms13033959
43. Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi:10.1126/science.1141478
44. Liang Y, Zhao J, Zou H, Zhang J, Zhang T. In vitro and in silico evaluation of EGFR targeting activities of curcumin and its derivatives. Food Funct. 2021;12(21):10667–10675. doi:10.1039/D1FO02002A
45. Ahsan MJ, Khalilullah H, Yasmin S, Jadav SS, Govindasamy J. Synthesis, characterisation, and in vitro anticancer activity of curcumin analogues bearing pyrazole/pyrimidine ring targeting EGFR tyrosine kinase. Biomed Res Int. 2013;2013:239354. doi:10.1155/2013/239354
46. Jiang AP, Zhou DH, Meng XL, et al. Down-regulation of epidermal growth factor receptor by curcumin-induced UBE1L in human bronchial epithelial cells. J Nutr Biochem. 2014;25(2):241–249. doi:10.1016/j.jnutbio.2013.11.001
47. van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65(23):3756–3788. doi:10.1007/s00018-008-8281-1
48. Shiozaki H, Oka H, Inoue M, Tamura S, Monden M. E-cadherin mediated adhesion system in cancer cells. Cancer. 1996;77(8 Suppl):1605–1613.
49. Rodríguez Castaño P, Parween S, Pandey AV. Bioactivity of curcumin on the cytochrome P450 enzymes of the steroidogenic pathway. Int J Mol Sci. 2019;20(18):4606. doi:10.3390/ijms20184606
50. Chen HW, Lee JY, Huang JY, et al. Curcumin inhibits lung cancer cell invasion and metastasis through the tumor suppressor HLJ1. Cancer Res. 2008;68(18):7428–7438. doi:10.1158/0008-5472.CAN-07-6734
51. Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi:10.1080/22221751.2020.1719902
52. Zhang Q, Yu H, Qi J, et al. Natural formulas and the nature of formulas: exploring potential therapeutic targets based on traditional Chinese herbal formulas. PLoS One. 2017;12(2):e0171628. doi:10.1371/journal.pone.0171628
53. Morris GM, Lim-Wilby M. Molecular docking. Methods Mol Biol. 2008;443:365–382.
54. Saikia S, Bordoloi M. Molecular docking: challenges, advances and its use in drug discovery perspective. Curr Drug Targets. 2019;20(5):501–521. doi:10.2174/1389450119666181022153016
55. Matyszewski A, Czarnecka A, Kawecki M, et al. Impaired glucose metabolism treatment and carcinogenesis. Oncol Lett. 2015;10(2):589–594. doi:10.3892/ol.2015.3324
56. Matyszewski A, Czarnecka AM, Solarek W, et al. Molecular basis of carcinogenesis in diabetic patients (review). Int J Oncol. 2015;46(4):1435–1443. doi:10.3892/ijo.2015.2865
57. Reuveni H, Flashner-Abramson E, Steiner L, et al. Therapeutic destruction of insulin receptor substrates for cancer treatment. Cancer Res. 2013;73(14):4383–4394. doi:10.1158/0008-5472.CAN-12-3385
58. Wang G, Yin L, Peng Y, et al. Insulin promotes invasion and migration of KRAS(G12D) mutant HPNE cells by upregulating MMP-2 gelatinolytic activity via ERK- and PI3K-dependent signalling. Cell Prolif. 2019;52(3):e12575. doi:10.1111/cpr.12575
59. Sarkar PL, Lee W, Williams ED, et al. Nelson CC: insulin enhances migration and invasion in prostate cancer cells by up-regulation of FOXC2. Front Endocrinol (Lausanne). 2019;10:481. doi:10.3389/fendo.2019.00481
60. Arcidiacono B, Iiritano S, Nocera A, et al. Insulin resistance and cancer risk: an overview of the pathogenetic mechanisms. Exp Diabetes Res. 2012;2012:789174. doi:10.1155/2012/789174
61. Argilés JM, López-Soriano FJ. Insulin and cancer (Review). Int J Oncol. 2001;18(4):683–687.
62. Parekh N, Guffanti G, Lin Y, Ochs-Balcom HM, Makarem N, Hayes R. Insulin receptor variants and obesity-related cancers in the Framingham Heart Study. Cancer Causes Control. 2015;26(8):1189–1195. doi:10.1007/s10552-015-0613-5
63. Janssen P, Rotondo A, Mulé F, Tack J. Review article: a comparison of glucagon-like peptides 1 and 2. Aliment Pharmacol Ther. 2013;37(1):18–36. doi:10.1111/apt.12092
64. Campbell JE, Drucker DJ. Islet α cells and glucagon–critical regulators of energy homeostasis. Nat Rev Endocrinol. 2015;11(6):329–338. doi:10.1038/nrendo.2015.51
65. Hancock AS, Du A, Liu J, Miller M, May CL. Glucagon deficiency reduces hepatic glucose production and improves glucose tolerance in adult mice. Mol Endocrinol. 2010;24(8):1605–1614. doi:10.1210/me.2010-0120
66. Moyer MP, Aust JB, Dixon PS, Levine BA, Sirinek KR. Glucagon enhances growth of cultured human colorectal cancer cells in vitro. Am J Surg. 1985;150(6):676–679. doi:10.1016/0002-9610(85)90406-4
67. Yagi T, Kubota E, Koyama H, et al. Glucagon promotes colon cancer cell growth via regulating AMPK and MAPK pathways. Oncotarget. 2018;9(12):10650–10664. doi:10.18632/oncotarget.24367
68. Adeva-Andany MM, Funcasta-Calderón R, Fernández-Fernández C, Castro-Quintela E, Carneiro-Freire N. Metabolic effects of glucagon in humans. J Clin Transl Endocrinol. 2019;15:45–53. doi:10.1016/j.jcte.2018.12.005
69. Vivacqua A, De Marco P, Belfiore A, Maggiolini M. Recent advances on the role of microRNAs in both insulin resistance and cancer. Curr Pharm Des. 2017;23(25):3658–3666. doi:10.2174/1381612823666170622105123
70. Vigneri R, Goldfine ID, Frittitta L. Insulin, insulin receptors, and cancer. J Endocrinol Invest. 2016;39(12):1365–1376. doi:10.1007/s40618-016-0508-7
71. Weichhaus M, Broom J, Wahle K, Bermano G. A novel role for insulin resistance in the connection between obesity and postmenopausal breast cancer. Int J Oncol. 2012;41(2):745–752. doi:10.3892/ijo.2012.1480
72. Cohen DH, LeRoith D. Obesity, type 2 diabetes, and cancer: the insulin and IGF connection. Endocr Relat Cancer. 2012;19(5):F27–45. doi:10.1530/ERC-11-0374
73. Gunter MJ, Xie X, Xue X, et al. Breast cancer risk in metabolically healthy but overweight postmenopausal women. Cancer Res. 2015;75(2):270–274. doi:10.1158/0008-5472.CAN-14-2317
74. Hernandez AV, Guarnizo M, Miranda Y, et al. Association between insulin resistance and breast carcinoma: a systematic review and meta-analysis. PLoS One. 2014;9(6):e99317. doi:10.1371/journal.pone.0099317
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.