Back to Journals » Journal of Inflammation Research » Volume 16
Identifying Immune Cell Infiltration and Hub Genes During the Myocardial Remodeling Process After Myocardial Infarction
Authors Tian Y, Wang Z, Liang F, Wang Y
Received 12 April 2023
Accepted for publication 28 June 2023
Published 11 July 2023 Volume 2023:16 Pages 2893—2906
DOI https://doi.org/10.2147/JIR.S416914
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Ning Quan
Yuan Tian,1,* Zilin Wang,2,* Feng Liang,3,* Yi Wang1
1Department of Cardiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200080, People’s Republic of China; 2Department of Radiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200080, People’s Republic of China; 3Heart Center, Shanghai Chest Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200080, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yi Wang, Department of Cardiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, NO. 100 Haining Road, Shanghai, 200080, People’s Republic of China, Tel/Fax +86 021 63241377, Email [email protected]
Purpose: Myocardial remodeling after myocardial infarction (MI) is a complex repair process following myocardial injury, characterized by the infiltration of multiple types of immune cells. However, the underlying molecular mechanism of myocardial remodeling after MI remains obscure. This study aimed to identify the hub differential expression genes (DEGs) of myocardial remodeling after MI and determine the distribution of immune cells infiltrating the pathology.
Methods: We downloaded GSE132143, GSE151834, and GSE176092 data from the GEO database. The GSE132143 dataset was used to identify DEGs, perform functional annotation, and screen hub genes based on protein-protein interaction (PPI) analysis. The GSE151834 dataset was used to validate the expression of hub genes. CIBERSORTx analysis was performed to explore the immune microenvironment in myocardial remodeling after MI. After conducting a literature review, we selected P3H3 to confirm the expression by utilizing immunohistochemistry and qRT-PCR. Finally, the snRNA-seq data in dataset GSE176092 was used for clarifying the expression of these hub genes in various cell clusters.
Results: We found 975 DEGs in myocardial remodeling after MI. Four hub genes (P3H3, COL15A1, COL16A1, COL27A1) were identified and were verified in the GSE151834 dataset. According to immune infiltration analysis, CD4+ naive T cells, regulatory T cells, monocytes, M2 macrophages, and neutrophils were involved in the pathological process of myocardial remodeling after MI. Additionally, in vitro experiments verified that P3h3 expression was significantly elevated in myocardial remodeling after MI. The snRNA-seq data analyzed that P3h3, Col15a1, Col16a1, and Col27a1 were highly expressed in fibroblasts of post-MI.
Conclusion: This study identified four hub genes P3H3, COL15A1, COL16A1, and COL27A1, particularly P3H3, as potential targets for targeted therapy in MI patients.
Keywords: myocardial infarction, myocardial remodeling, hub genes, immune infiltration, bioinformatic analysis
Introduction
Acute myocardial infarction (MI) is the leading cause of global morbidity and mortality. About 30% of MI patients undergo myocardial remodeling after MI, and these patients have a higher risk of mortality.1 The myocardial remodeling after MI is a complex reparative process with 3 overlapping phases: the inflammatory phase, proliferative phase, and healing phase.2 In the early phase of infarction, the inflammatory response is activated, and immune cells then accumulate in the ischemic infarct area to participate in the inflammatory cascade.3 Meanwhile, resident cardiac fibroblasts are activated by early tissue damage.4 During the proliferative and healing phases, these cells proliferate, migrate into wounded tissue, and only differentiate into myofibroblasts, which initiate and promote scar formation and fibrosis.4 Moderate inflammation response helps cardiac repair, whereas excessive inflammation worsens cardiac injury and post-MI myocardial remodeling.5 Current treatments for MI consist solely of cause-dependent intervention, which is incapable of minimizing myocardial necrosis and maximizing post-MI heart healing. In addition to revascularization, new treatment strategies should consider the molecular mechanisms that regulate post-MI myocardial remodeling.6 The discovery of new collagen genes, essential plasma proteins, and a novel macrophage phenotype has enhanced our understanding of the molecular mechanisms underlying post-MI myocardial remodeling in humans.7–9 Nevertheless, the underlying mechanisms of post-MI myocardial remodeling remain unclear.
Over the years, bioinformatic analysis based on RNA-sequencing (RNA-seq) data has been increasingly applied to the identification of the key genes, yielding crucial insights into underlying mechanisms of disease progression.10–12 Moreover, CIBERSORTx, a computational algorithm for characterizing relative fractions of diverse cell subsets of gene expression profiles,13 is increasingly being used to estimate the immune cell composition of complex tissues from their RNA-seq data.14–16 However, RNA profiling is limited to ensemble-based approaches that cannot provide biological insights into specific cell types.17 With the emergence and development of single-cell techniques, researchers can now explore the function and role of distinct cell types in disease progression.17,18 Nevertheless, the number of detected genes based on single-cell techniques is significantly lower than that of bulk RNA-seq.18 Thus, the integration of RNA-seq and single-cell techniques is essential for elucidating the biomarkers and molecular mechanisms underlying the development of disease.17
To investigate hub genes and immune cell landscape during myocardial remodeling after MI, we performed a comprehensive analysis of RNA-seq data of human post-MI myocardial remodeling, which identified differential expression genes (DEGs), explored the underlying biological functions and pathways, and further screened hub genes by constructing protein-protein interaction (PPI) network. The expression of hub genes was then validated in the RNA-seq data of post-MI mice. Next, we used CIBERSORTx to determine the percentage of immune infiltration of human post-MI myocardial remodeling and performed immune infiltration correlation analyses of hub genes. Furthermore, our candidate gene P3H3 was further validated using immunohistochemistry (ICH) and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR). Finally, single nuclei RNA sequencing (snRNA-seq) data of post-MI mice was used to determine the expression and distribution of hub genes. The workflow of this study is shown in Figure 1.
 |
Figure 1 Workflow chart of the study. |
Materials and Methods
Data Acquisition
Two RNA-seq datasets (GSE132143 and GSE151834) and an snRNA-seq dataset (GSE176092) were acquired from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The GSE132143 dataset contained 20 post-MI myocardial remodeling samples and 12 normal samples from the human ventricle. The GSE151834 dataset, containing 4 injured tissues and 4 uninjured remote tissues from 4 post-MI (2 weeks) mice ventricles, was used as an independent validation set. The snRNA-seq dataset GSE176092, containing filtered cardiomyocytes of 2 injured tissues and 2 uninjured remote tissues from 2 post-MI (2 weeks) mice ventricles, was used to clarify the expression and distribution of hub genes. The uninjured remote tissues from post-MI mice can be considered as the control samples of infarcted tissues.19
Processing of the Datasets and DEGs Screening
The dataset GSE132143 was translated into the gene symbol by the R package “org.Hs.eg.db”. Then, it was log2 transformed and quantile normalized by the “normalizeBetweenArrays” function of the “limma” R package.20,21 Principal component analysis (PCA) was calculated using “prcomp” function to verify the repeatability of samples. Next, DEGs in the post-MI myocardial remodeling were screened by using the “limma” R package21 with cut-off criteria of the false-discovery rate (FDR, Benjamini-Hochberg correction method) < 0.05 and |log fold change (FC)| > 1.5.
Functional Enrichment Analysis
Gene ontology and signaling pathways associated with DEGs have a significant impact on the development of post-MI myocardial remodeling. GO analysis and KEGG pathway analysis were performed by the R package “clusterProfiler”.21 FDR < 0.05 was considered statistically significant.
Identification of Protein-Protein Interaction (PPI) Networks and Hub Genes
The construction and visualization of the PPI network based on DEGs were carried out with the online STRING database (http://string-db.org/).22 The combined score > 0.4 was set as the threshold value. Next, the result of the PPI network was further imported into Cytoscape software (version 3.10.0) to identify critical subnetworks and hub genes through the plug-ins in Cytoscape.23 Specifically, Molecular Complexity Detection (MCODE),24 a plugin in Cytoscape, was utilized to identify the most important modules based on the PPI network. Degree Cutoff = 2, Node Score Cutoff = 0.2, K-core = 2, Max. depth = 100 and MCODE scores ≥10 were set as filtering parameters. Furthermore, CytoHubba,25 a plugin in Cytoscape, was utilized to calculate the top 10 core genes of the PPI network based on Maximal Clique Centrality (MCC) and Density of Maximum Neighborhood Component (DMNC) algorithms. The hub genes were determined by taking the overlapping genes of MCODE (significant module 1), MCC, and DMNC algorithms.
The Accuracy of the Hub Genes
The ROC curves were generated by using the “pROC” package,26 while the area under the curve (AUC) was calculated to evaluate the predictive accuracy of the hub genes for myocardial remodeling after MI. An AUC > 0.9 indicated that the hub genes had a good fitting effect.
Immune Cell Landscapes and Immune Correlation Analysis
The CIBERSORTx13 (https://cibersortx.stanford.edu/) was used to analyze the fractions of infiltrating immune cells in the GSE132143 samples with gene signature file LM22 set at 100 permutations. Spearman correlation analysis performed by “corrplot” package was used to evaluate the association of immune cells with each other. The associations of the hub genes with infiltrating immune cells were evaluated using Pairwise Spearman correlation analysis with family-wise error rate (FWER, Holm correction method). The resulting associations were visualized using the “ggplot2” package. FWER < 0.05 was considered statistically significant.
Animal Models
The infarct tissue from 2 weeks post-MI mice ventricular was defined as being in a late stage of myocardial remodeling.27 Eight-week-old male C57BL/6 mice used in this study were purchased from SiPeiFu (Beijing, China) and randomly assigned to the MI (n=6) or sham group (n=4). As previously described, the MI model was constructed by permanent ligation of the left anterior descending (LAD) artery.28 After chest hair removal, rodents were placed in an anesthesia box for rapid isoflurane (2%) anesthesia and then placed supine on the experimental plate. Following a left-sided thoracotomy, the heart was swiftly removed. The LAD artery was ligated 2 mm from its origin using a silk suture of size 6–0. After returning to the heart and expelling the gas from the chest cavity, the chest was closed with a 5–0 silk suture. The sham group received the same surgical procedure, but coronary artery ligation was not performed. All mice were housed in an environment with a light/dark cycle of 12/12 h and had free access to food and water. All animal experiments were approved by the Institutional Animal Care and Use Committee of Shanghai General Hospital (NO.2023AW004) and followed the standards of the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Echocardiography
Immunohistochemical Analysis
qRT-PCR Assay of the P3h3
SnRNA-Seq Data Analysis
Statistical Analysis
The data are expressed as the mean ± SD. Unpaired Student’s T-test and Mann–Whitney U-test were applied for comparisons between the two groups. All statistical analyses were performed via R software 4.2.0 and GraphPad Prism 9. P value < 0.05 was considered statistically significant.
Results
Identification of DEGs in Myocardial Remodeling After MI
To determine any significant differences in the gene expression profile among samples in GSE132143, we analyzed data distribution before and after quantile normalization through box plots. After normalization, the differences among samples were eliminated (Supplementary Figure 1A and B). We further validated the interpretability and scalability of the data based on PCA dimension reduction. PCA demonstrated that post-MI myocardial remodeling samples clustered together (Figure 2A). Principal component-1 (PC1) separated the transcriptional fingerprints of the two groups, suggesting that there were differences in gene expression between the two groups (Figure 2A). A total of 566 up-regulated DEGs and 409 down-regulated DEGs were identified (Figure 2B). The heatmap was generated to visualize the DEGs in the meta-data cohort (Figure 2C).
Functional Enrichment Analysis
To investigate the potential molecular mechanisms and pathways of post-MI cardiac remodeling, we performed GO and KEGG analyses on DEGs. GO enrichment analysis revealed that DEGs in the biological process (BP) were mainly enriched in the cellular divalent inorganic cation homeostasis, modulation of chemical synaptic transmission, regulation of trans-synaptic signaling, extracellular matrix organization, extracellular structure organization, external encapsulating structure organization (Figure 3A). The results of cellular component (CC) were significantly enriched in the collagen-containing extracellular matrix, presynapse, postsynaptic specialization, and neuron-to-neuron synapse (Figure 3A). As for molecular function (MF), the DEGs were mainly focused on extracellular matrix structural constituents, glycosaminoglycan binding, and immune receptor activity (Figure 3A). KEGG analysis revealed that the DEGs were mainly enriched in pathways of neutrophil extracellular trap (NET) formation, protein digestion and absorption, phagosome, and extracellular matrix (ECM) receptor interaction (Figure 3B).
 |
Figure 3 Functional enrichment analysis of DEGs. (A) GO analysis on DEGs. (B) KEGG analysis on DEGs. |
Identification and Validation of Hub Gene
A total of 975 DEGs were used to construct a PPI network using the STRING online platform. There were 680 nodes and 4740 edges (Figure 4A). To screen the hub genes, the critical subnetworks were extracted with an MCODE plug-in based on the PPI network. Three significant modules (subnetworks) closely related to the PPI network were found and separated (Supplementary Table 1). The most significant module 1 composed of 14 nodes (Figure 4B). Then, the Cytoscape software was also used to analyze the PPI network. The critical subnetworks and hub genes of MCC and DMNC topological algorithms are shown in Figures 4C, D and Supplementary Table 2. Among the results of MCODE (significant module 1), MCC, and DMNC algorithms, the overlapping genes were as follows: P3H3, COL15A1, COL16A1, and COL27A1 (Figure 4E). Furthermore, to demonstrate the accuracy of these hub genes, the dataset GSE151834 was used for external verification. The results revealed that the expression levels of P3h3, Col15a1, Col16a1, and Col27a1 were significantly higher in the injured group than in the uninjured remote group (Figure 5). These 4 hub genes may serve as significant biomarkers in the myocardial remodeling process.
The Accuracy of Hub Genes in Myocardial Remodeling After MI
The diagnostic ability of the four hub genes to distinguish post-MI myocardial remodeling samples from the normal samples exhibited a favorable fitting effect, with AUCs of 1.000, 0.946, 0.996, and 1.000 for P3H3, COL16A1, COL15A1, and COL27A1 in the meta-data cohort, respectively (Figure 6A). Moreover, to confirm the discrimination ability of those hub genes, the AUCs of all four hub genes, P3h3, Col16a1, Col15a1, and Col27a1, were 1.000 in the GSE151834 dataset (Figure 6B).
Immune Cell Landscapes and Immune Correlation Analysis
The immune microenvironment plays an important role in myocardial remodeling after MI.3 To further explore the relationship between hub genes and immune cells, we studied the subpopulations of infiltrated immune cells in post-MI myocardial remodeling and normal samples. The abundance distribution of 22 infiltrated immune cell types in each sample was presented in a histogram (Supplementary Figure 2A) and heatmap (Supplementary Figure 2B). Post-MI myocardial remodeling samples had significantly higher proportions of CD4+ naive T cells (p < 0.05), regulatory T cells (p < 0.05), and monocytes (p < 0.01) than normal samples. The proportions of M2 macrophages (p < 0.0001) and neutrophils (p < 0.01) were significantly lower in post-MI myocardial remodeling samples than in normal samples (Figure 7A). Interestingly, both post-MI myocardial remodeling and normal samples lacked activated mast cells, CD4+ memory-activated T cells, and gamma delta T cells. Furthermore, both P3H3 and COL16A1 were positively correlated with Regulatory T cells (FWER < 0.05) and monocytes (FWER < 0.05), whereas they were negatively correlated with M2 macrophages (both FWER < 0.01) (Figure 7B). The M2 macrophages showed a significantly negative correlation with monocytes (r = −0.6, p < 0.001) (Figure 7C).
Overexpression of P3h3 in Post-MI Myocardial Remodeling Samples
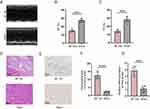
After reviewing the literature, three hub genes including Col15a1, Col16a1, and Col27a1 have been reported to increase in infarcted tissue from 14-day post-MI mice via qRT-PCR.19 Therefore, we choose to validate the expression of P3h3 by IHC and qRT-PCR analyses. Figure 8A depicts echocardiographic images representative of the MI and sham samples. The cardiac function confirmed by echocardiography showed a significant reduction in contractility, as reflected by left ventricle EF (29.7 ± 3.9% versus 55.0 ± 4.6%, p < 0.001) and FS (14.0 ± 2.0% versus 28.3 ± 3.0%, p < 0.001) in the MI group compared with that in the sham group (Figure 8B and C). As depicted in Figure 8D, the cardiac tissue in the MI group showed significant myocardial infarction, but not in the sham group. The positive area of P3h3 in the MI group was significantly higher (p < 0.0001) than that in the sham group (Figure 8E and F). Moreover, the P3h3 mRNA expression level in the MI samples was significantly higher (p < 0.001) than that in the sham samples (Figure 8G).
Expression Prediction of Hub Genes in snRNA-Seq Profiling
To elucidate the expression and distribution of hub genes in post-MI myocardial remodeling, we analyzed the data of filtered cardiomyocytes from a public snRNA database. According to the UMAP algorithm, cells were successfully classified into 6 separate clusters (Figure 9A), which include cardiomyocytes (Ryr2), endothelial cells (Pecam1), fibroblasts (Dcn), pericardial cells (Upk3b), myeloid cells (C1qa), and pericytes (Rgs5) (Supplementary Figure 3). The marker genes in 6 cell clusters are listed in Supplementary Figure 3. We found that the P3h3, Col16a1, Col15a1, and Col27a1 were highly expressed in fibroblasts of post-MI myocardial remodeling (Figure 9B). Moreover, P3h3 and Col16a1 were highly expressed in pericytes and pericardial cells of post-MI myocardial remodeling, respectively (Figure 9B).
Discussion
Although significant progress has been made in the treatment of MI, strategies to restore damaged tissue and reconstruct organ function are still required. Therefore, it is necessary to explore the specific mechanisms of post-MI myocardial remodeling to motivate the development of new therapeutic strategies and thereby improve the prognosis of patients with MI. In this study, we identified hub genes and the landscape of immune infiltration in human post-MI myocardial remodeling.
We first identified 975 DEGs of post-MI myocardial remodeling by bioinformatics analysis. The potential biological functions of these DEGs were investigated by GO and KEGG enrichment analyses, which mainly indicated the key roles of inflammatory response and extracellular matrix. The dynamic alterations in ECM composition may contribute to cardiac repair following MI.29 Some approaches that strictly regulate ECM deposition and crosslinking may prevent adverse remodeling and heart failure after MI.30
Subsequently, among the significant DEGs, we identified four hub genes for post-MI cardiac remodeling, including P3H3, COL15A1, COL16A1, and COL27A1, and validated them using the GSE151834 dataset. COL15A1 is essential for the organization of the extracellular matrix and structural and functional microvessels in the heart that can promote myocardial proliferation.31,32 It can modulate the elasticity of the myocardium, and its deficiency leads to left ventricular dilatation and left ventricular wall thinning.31 COL16A1 is a linker protein that contributes to the organization of large fiber networks, thereby regulating the integrity and stability of the ECM. It is positively associated with left ventricular dysfunction in patients with ischemic cardiomyopathy.7 COL27A1 is known to be regulated by SOX9.33 The role of COL27A1 in post-MI myocardial remodeling requires additional study. P3H3, a member of the prolyl 3-hydroxylase family, is involved in collagen post-translational modifications. Database analyses of vertebrate genomes showed that the prolyl 3-hydroxylase family had three isoenzymes named P3H1, P3H2, and P3H3.34 Although the prolyl 3-hydroxylase family was discovered in collagens more than half a century ago, its exact functions remain unclear.35,36 P3h3 has been reported to form a complex with Plod1 and P3h4 that is necessary for the normal assembly and cross-linking of collagen fibrils.37 P3h3−/− mice displayed severe under-hydroxylation of lysines, especially at cross-linking sites in type I collagen from the bone, cornea, and aorta.38
In addition, we observed significant differences in the abundance of infiltrating immune cell subpopulations between the two groups, emphasizing the role of immune cells in the development of post-MI myocardial remodeling. In our study, the proportion of CD4+ naive T cells,39 regulatory T cells, and monocytes40 increased, whereas the proportion of M2 macrophages and neutrophils decreased. It has been demonstrated that regulatory T cells in the post-MI-damaged myocardium enhance cardiac function, promote scar repair, and inhibit ventricular remodeling.41,42 CD4+ T cells become activated after MI, thus promoting wound healing and scar formation.43 Nonetheless, persistently increased CD4+ T cells regulated by TNFR1/ TNFR2 promote ventricular remodeling and progressive cardiac dysfunction during ischemic heart failure.39,44 Moreover, neutrophils can promote M2 macrophage polarization.45 M2 macrophages can produce multiple anti-inflammatories, proangiogenic, and pro-reparative factors, as well as engulf and clear dead cells, thereby enhancing infarct healing and reducing adverse cardiac remodeling following acute MI.46 Our bioinformatics analysis highlights the importance of these cells in the pathogenesis of myocardial remodeling after MI in humans.
Given that the expression of P3H3 has not been described in other studies, we further verified the elevated expression of P3h3 using immunohistochemical and qRT-PCR analyses. Additionally, snRNA-seq data analysis revealed that P3h3, Col15a1, Col16a1, and Col27a1 were highly expressed in fibroblasts in post-MI myocardial remodeling, among which COL15A1 is expressed in fibroblasts of the human heart.32,47 A recent study showed that P3H3 may be associated with cardiac fibroblast activation mediated by transforming growth factor beta.48 We hypothesize that P3H3 plays a crucial role in the assembly mechanism and molecular packing of collagen fibrils following cardiac injury. However, this may require further in vitro experiment validation.
This research has several limitations. The precise mechanisms of P3H3, COL15A1, COL16A1, and COL27A1 on the development of myocardial remodeling after MI remain unclear and require further investigation. In addition, the expression and distribution of these genes in subpopulations of immune cells have yet to be determined.
In this study, we assess the immune cell landscape in post-MI myocardial remodeling by using RNAseq data from human heart tissue, and identify P3H3 expression after MI. These findings increase our understanding of the role of immuno-microenvironment in cardiac injury repair and emphasize the significance of collagen post-translational modifications in myocardial remodeling. However, the impact of the prolyl 3-hydroxylase family on post-MI myocardial remodeling is unknown and should be the subject of future research.
Conclusion
Our study identified four hub genes, including P3H3, COL15A1, COL16A1, and COL27A1, as well as the immune cell landscapes in post-MI myocardial remodeling. These findings may contribute to a greater understanding of the mechanism underlying the development of myocardial remodeling after MI and provide novel ideas for the drug treatment of post-MI myocardial remodeling.
Data Sharing Statement
The data of bioinformatics analysis (GSE132143, GSE151834, and GSE176092) for this study can be found in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Further inquiries can be directed to the corresponding author.
Ethics Statement
All animal experiments were approved by the Institutional Animal Care and Use Committee of Shanghai General Hospital (NO.2023AW004) and followed the standards of the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Acknowledgments
The authors would like to thank Dr. Win Topatana from Zhejiang University for the revision of this manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81470471), and the Natural Science Foundation of Shanghai (No. 19ZR11440600).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Curley D, Lavin Plaza B, Shah AM, Botnar RM. Molecular imaging of cardiac remodelling after myocardial infarction. Basic Res Cardiol. 2018;113(2):10. doi:10.1007/s00395-018-0668-z
2. Frangogiannis NG. The inflammatory response in myocardial injury, repair, and remodelling. Nat Rev Cardiol. 2014;11(5):255–265. doi:10.1038/nrcardio.2014.28
3. Swirski FK, Nahrendorf M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol. 2018;18(12):733–744. doi:10.1038/s41577-018-0065-8
4. Burke RM, Burgos Villar KN, Small EM. Fibroblast contributions to ischemic cardiac remodeling. Cell Signal. 2021;77:109824. doi:10.1016/j.cellsig.2020.109824
5. Sun K, Li YY, Jin J. A double-edged sword of immuno-microenvironment in cardiac homeostasis and injury repair. Signal Transduct Target Ther. 2021;6(1):79. doi:10.1038/s41392-020-00455-6
6. Frantz S, Hundertmark MJ, Schulz-Menger J, Bengel FM, Bauersachs J. Left ventricular remodelling post-myocardial infarction: pathophysiology, imaging, and novel therapies. Eur Heart J. 2022;43(27):2549–2561. doi:10.1093/eurheartj/ehac223
7. Gil-Cayuela C, Rivera M, Ortega A, et al. RNA sequencing analysis identifies new human collagen genes involved in cardiac remodeling. J Am Coll Cardiol. 2015;65(12):1265–1267. doi:10.1016/j.jacc.2015.01.029
8. Chan MY, Efthymios M, Tan SH, et al. Prioritizing Candidates of Post-Myocardial Infarction Heart Failure Using Plasma Proteomics and Single-Cell Transcriptomics. Circulation. 2020;142(15):1408–1421. doi:10.1161/CIRCULATIONAHA.119.045158
9. Deniset JF, Belke D, Lee WY, et al. Gata6(+) Pericardial Cavity Macrophages Relocate to the Injured Heart and Prevent Cardiac Fibrosis. Immunity. 2019;51(1):131–140 e5. doi:10.1016/j.immuni.2019.06.010
10. Zhao J, Lv T, Quan J, et al. Identification of target genes in cardiomyopathy with fibrosis and cardiac remodeling. J Biomed Sci. 2018;25(1):63. doi:10.1186/s12929-018-0459-8
11. Rogers JD, Aguado BA, Watts KM, Anseth KS, Richardson WJ. Network modeling predicts personalized gene expression and drug responses in valve myofibroblasts cultured with patient sera. Proc Natl Acad Sci U S A. 2022;119(8). doi:10.1073/pnas.2117323119
12. Auwul MR, Rahman MR, Gov E, Shahjaman M, Moni MA. Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19. Brief Bioinform. 2021;22(5). doi:10.1093/bib/bbab120
13. Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi:10.1038/s41587-019-0114-2
14. Casarrubios M, Provencio M, Nadal E, et al. Tumor microenvironment gene expression profiles associated to complete pathological response and disease progression in resectable NSCLC patients treated with neoadjuvant chemoimmunotherapy. J Immunother Cancer. 2022;10(9). doi:10.1136/jitc-2022-005320
15. Nevalainen T, Autio A, Hurme M. Composition of the infiltrating immune cells in the brain of healthy individuals: effect of aging. Immun Ageing. 2022;19(1):45. doi:10.1186/s12979-022-00302-y
16. Innocenti F, Yazdani A, Rashid N, et al. Tumor Immunogenomic Features Determine Outcomes in Patients with Metastatic Colorectal Cancer Treated with Standard-of-Care Combinations of Bevacizumab and Cetuximab. Clin Cancer Res. 2022;28(8):1690–1700. doi:10.1158/1078-0432.CCR-21-3202
17. Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16(3):133–145. doi:10.1038/nrg3833
18. Artyomov MN, Van den Bossche J. Immunometabolism in the Single-Cell Era. Cell Metab. 2020;32(5):710–725. doi:10.1016/j.cmet.2020.09.013
19. Yokota T, McCourt J, Ma F, et al. Type V Collagen in Scar Tissue Regulates the Size of Scar after Heart Injury. Cell. 2020;182(3):545–562 e23. doi:10.1016/j.cell.2020.06.030
20. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi:10.1093/bioinformatics/19.2.185
21. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
22. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–D646. doi:10.1093/nar/gkac1000
23. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
24. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi:10.1186/1471-2105-4-2
25. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
26. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi:10.1186/1471-2105-12-77
27. Forte E, Skelly DA, Chen M, et al. Dynamic Interstitial Cell Response during Myocardial Infarction Predicts Resilience to Rupture in Genetically Diverse Mice. Cell Rep. 2020;30(9):3149–3163 e6. doi:10.1016/j.celrep.2020.02.008
28. Wang Z, Qiu Z, Hua S, et al. Nuclear Tkt promotes ischemic heart failure via the cleaved Parp1/Aif axis. Basic Res Cardiol. 2022;117(1):18. doi:10.1007/s00395-022-00925-8
29. Dobaczewski M, Gonzalez-Quesada C, Frangogiannis NG. The extracellular matrix as a modulator of the inflammatory and reparative response following myocardial infarction. J Mol Cell Cardiol. 2010;48(3):504–511. doi:10.1016/j.yjmcc.2009.07.015
30. Frangogiannis NG. The extracellular matrix in myocardial injury, repair, and remodeling. J Clin Invest. 2017;127(5):1600–1612. doi:10.1172/JCI87491
31. Rasi K, Piuhola J, Czabanka M, et al. Collagen XV is necessary for modeling of the extracellular matrix and its deficiency predisposes to cardiomyopathy. Circ Res. 2010;107(10):1241–1252. doi:10.1161/CIRCRESAHA.110.222133
32. Rhee S, Paik DT, Yang JY, et al. Endocardial/endothelial angiocrines regulate cardiomyocyte development and maturation and induce features of ventricular non-compaction. Eur Heart J. 2021;42(41):4264–4276. doi:10.1093/eurheartj/ehab298
33. Jenkins E, Moss JB, Pace JM, Bridgewater LC. The new collagen gene COL27A1 contains SOX9-responsive enhancer elements. Matrix Biol. 2005;24(3):177–184. doi:10.1016/j.matbio.2005.02.004
34. Vranka JA, Sakai LY, Bachinger HP. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004;279(22):23615–23621. doi:10.1074/jbc.M312807200
35. Ogle JD, Arlinghaus RB, Lgan MA. 3-Hydroxyproline, a new amino acid of collagen. J Biol Chem. 1962;237:3667–3673.
36. Hudson DM, Eyre DR. Collagen prolyl 3-hydroxylation: a major role for a minor post-translational modification? Connect Tissue Res. 2013;54(4–5):245–251. doi:10.3109/03008207.2013.800867
37. Heard ME, Besio R, Weis M, et al. Sc65-Null Mice Provide Evidence for a Novel Endoplasmic Reticulum Complex Regulating Collagen Lysyl Hydroxylation. PLoS Genet. 2016;12(4):e1006002. doi:10.1371/journal.pgen.1006002
38. Hudson DM, Weis M, Rai J, et al. P3h3-null and Sc65-null Mice Phenocopy the Collagen Lysine Under-hydroxylation and Cross-linking Abnormality of Ehlers-Danlos Syndrome Type VIA. J Biol Chem. 2017;292(9):3877–3887. doi:10.1074/jbc.M116.762245
39. Kumar V, Prabhu SD, Bansal SS. CD4(+) T-lymphocytes exhibit biphasic kinetics post-myocardial infarction. Front Cardiovasc Med. 2022;9:992653. doi:10.3389/fcvm.2022.992653
40. van der Laan AM, Ter Horst EN, Delewi R, et al. Monocyte subset accumulation in the human heart following acute myocardial infarction and the role of the spleen as monocyte reservoir. Eur Heart J. 2014;35(6):376–385. doi:10.1093/eurheartj/eht331
41. Tang TT, Yuan J, Zhu ZF, et al. Regulatory T cells ameliorate cardiac remodeling after myocardial infarction. Basic Res Cardiol. 2012;107(1):232. doi:10.1007/s00395-011-0232-6
42. Xia N, Lu Y, Gu M, et al. A Unique Population of Regulatory T Cells in Heart Potentiates Cardiac Protection From Myocardial Infarction. Circulation. 2020;142(20):1956–1973. doi:10.1161/CIRCULATIONAHA.120.046789
43. Hofmann U, Beyersdorf N, Weirather J, et al. Activation of CD4+ T lymphocytes improves wound healing and survival after experimental myocardial infarction in mice. Circulation. 2012;125(13):1652–1663. doi:10.1161/CIRCULATIONAHA.111.044164
44. Kumar V, Rosenzweig R, Asalla S, Nehra S, Prabhu SD, Bansal SS. TNFR1 Contributes to Activation-Induced Cell Death of Pathological CD4(+) T Lymphocytes During Ischemic Heart Failure. JACC Basic Transl Sci. 2022;7(10):1038–1049. doi:10.1016/j.jacbts.2022.05.005
45. Horckmans M, Ring L, Duchene J, et al. Neutrophils orchestrate post-myocardial infarction healing by polarizing macrophages towards a reparative phenotype. Eur Heart J. 2017;38(3):187–197. doi:10.1093/eurheartj/ehw002
46. ter Horst EN, Hakimzadeh N, van der Laan AM, Krijnen PA, Niessen HW, Piek JJ. Modulators of Macrophage Polarization Influence Healing of the Infarcted Myocardium. Int J Mol Sci. 2015;16(12):29583–29591. doi:10.3390/ijms161226187
47. Kivirikko S, Saarela J, Myers JC, Autio-Harmainen H, Pihlajaniemi T. Distribution of type XV collagen transcripts in human tissue and their production by muscle cells and fibroblasts. Am J Pathol. 1995;147(5):1500–1509.
48. Lozano J, Rai A, Lees JG, et al. Scalable Generation of Nanovesicles from Human-Induced Pluripotent Stem Cells for Cardiac Repair. Int J Mol Sci. 2022;23(22). doi:10.3390/ijms232214334
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.