Back to Journals » International Journal of General Medicine » Volume 15
Identification of the Level of Exosomal Protein by Parallel Reaction Monitoring Technology in HCC Patients
Authors Huang H, Zhang Q, Zhang Y, Sun X, Liu C, Wang Q, Huang Y, Li Q, Wu Z, Pu C, Sun A
Received 28 July 2022
Accepted for publication 5 October 2022
Published 14 October 2022 Volume 2022:15 Pages 7831—7842
DOI https://doi.org/10.2147/IJGM.S384140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Hui Huang,1 Qiqi Zhang,1 Yong Zhang,1 Xueying Sun,1 Chunyan Liu,1 Qi Wang,1 Yushuang Huang,1 Qingwei Li,2 Zepan Wu,2 Chunwen Pu,1 Aijun Sun1
1Department of Biobank, Dalian Public Health Clinical Center, Dalian, 116001, People’s Republic of China; 2College of Life Science, Liaoning Normal University, Dalian, 116081, People’s Republic of China
Correspondence: Chunwen Pu, Department of Biobank, Dalian Public Health Clinical Center, No. 269, Guibai Road, Ganjingzi District, Dalian, 116001, People’s Republic of China, Email [email protected] Aijun Sun, Department of biobank, Dalian Public Health Clinical Center, No. 269, Guibai Road, Ganjingzi District, Dalian, 116001, People’s Republic of China, Email [email protected]
Purpose: Reliable biomarkers for the diagnosis and differential diagnosis of various stages of liver cancer are lacking. In this study, we aim to detect the levels of differentially expressed proteins (DEPs) in serum exosomes of patients with different liver diseases using a sensitive method.
Patients and Methods: Exosomes were purified and validated. The expression of DEPs in exosomes from patients with chronic hepatitis B (CHB), liver cirrhosis (LC) and hepatocellular carcinoma (HCC) was validated by parallel reaction monitoring (PRM) technology and Western blotting, and the biological functions were analyzed by bioinformatics analysis.
Results: A total of 11 DEPs were identified by PRM technology. Significantly higher level of haptoglobin (Hp) was detected in HCC patients as compared to LC and CHB patients. HCC patients had a significantly lower level of transthyretin (TTR) in the patients with CHB. Among the patients with HCC who undertaken surgery, the postoperative levels of CRP, SERPINA3 and Heparin cofactor 2 (SERPIND1) were significantly reduced compared to their respective preoperative levels.
Conclusion: Hp and TTR may be potential markers for early diagnosis of HCC. CRP, SERPINA3 and SERPIND1 may serve as potential prognostic indicators for HCC patients undertaken surgery.
Keywords: exosome, hepatocellular carcinoma, mass spectrometry, biomarker
Introduction
As we all know, the diagnosis of liver disease is challenging, especially cirrhosis and liver cancer. Liver cancer can be detected by ultrasound, MRI, CT imaging or alpha-fetoprotein (AFP) of serum, accurate diagnosis is difficult due to lacking of obvious symptom during early development period. A new diagnostic method to distinguish the early stage of liver cancer have been trying to find, so as to improve the survival rate of patients.
In recent years, exosome as a new mechanism of intercellular communication has aroused widespread concern. The exosomes are membrane vesicle with a size of 30–100 nanometers,1 which are rich of lipids, proteins, RNAs and DNAs,1,2 mediating information transfer between cells. Studies have shown that exosomes are related to the occurrence and development of liver diseases, including hepatitis C virus (HCV) infection,3 hepatitis B virus (HBV) infection,4 hepatocellular carcinoma (HCC),5 liver fibrosis,6 cirrhosis,7 nonalcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD).8 In addition, almost all types of cells in the body can secrete exosomes, and exosomes can be widely detected in the human body, such as blood,9 urine,10 saliva,11 bile,12 milk,13 amniotic fluid14 and so on. Our previous study has found that the combined examination of extracellular miR-21 and miR-144 in serum is helpful in diagnosing HCC, and points out the enrichment of these two miRNAs in serum of patients with HCC,15 suggesting that extracellular vesicles or exosomes may provide new ideas for the diagnosis and prognosis of HCC.
Quantitative proteomics is one of the important applications in the field of proteomics research. Through quantitative proteomics technology, all proteins expressed in the same sample can be identified and quantified, DEPs between samples or groups can be screened, the function of differential proteins can be predicted combined with biological information analysis, and the physiological and pathological mechanisms can be analyzed. PRM technology is a targeted proteomics technology based on mass spectrometry and an ion monitoring technology based on high-resolution and high-precision mass spectrometry which can selectively detect the target protein and target peptide, so as to realize the quantification of the target protein/peptide. The PRM method is used to detect differential proteins and only quantitatively analyze the target protein. It is targeted, more accurate and more sensitive than other protein profiles. It is suitable for the verification of omics results and the quantitative analysis of specific proteins. Proteome analysis of serum exosomes can be used to identify diagnostic or prognostic biomarkers and provide in-depth understanding of disease development and progression mechanisms.
Quantitative differential proteomic analysis of serum exosomal proteins in patients with chronic hepatitis B (CHB), liver cirrhosis (LC) and hepatocellular carcinoma (HCC) were performed by using Label-free and iTRAQ proteomics technology in our previous study. In this study, we identified potential biomarkers and verified the expression of DEPs by PRM technology and Western blotting, and finally several biomarker candidate proteins were identified, aiming to play an important guiding role in the non-invasive diagnosis and prognosis of liver cancer.
Materials and Methods
Patient Characteristics
A total of 43 subjects from January 2016 to December 2018 were recruited from Dalian Public Health Clinical Center. The samples in this study were obtained with informed consent. The study was approved by Ethics Committee of the Dalian Public Health Clinical Center (Approval ID: 2020–032-001). Written informed consent was obtained from each participate before being enrolled into the study. Patient characteristics were shown in Tables 1 and 2. Recruits participating in the PRM mass spectrometry assay were divided into three groups showed in Table 1: CHB patients (n=10), LC patients (n=10), and HCC patients (n=10). Recruiters who participated in Western blotting protein validation were divided into three groups showed in Table 2: CHB patients (n=15), LC patients (n=15), and HCC patients (n=15). Some patients participated in both experiments. CHB, LC and HCC were diagnosed according to the criteria of the Asian Pacific Association for the Study of the Liver.
 |
Table 1 Patients Clinical Characteristics According to the Etiology in PRM Mass Spectrometry |
 |
Table 2 Patients Clinical Characteristics According to the Etiology in Western Blotting |
Purification of Human Serum Exosomes Using Exo-Spin Extracellular Vesicle (EV) Purification Kit
Serum exosomes of CHB, LC and HCC patients were extracted and purified using Exo-spin EV Purification Kit (Cell Guidance Systems, EX02). Cell debris was removed and exosomes were purified from 500 μL serum according to the Exo-spin EV Purification kit (Cell Guidance Systems, EX02) instructions.
Transmission Electron Microscopy and ZetaView Analysis
Serum exosomes were identified by transmission electron microscopy and ZetaView Analysis. The method in this part is consistent with our previous experimental method in identifying serum exosomes.15
Western Blotting Analysis
The cluster of differentiation 63 (CD63) and CD9 are often enriched in exosomes. To validate the isolation of the exosomes from the serums, Western blotting analysis was performed using the mouse polyclonal anti-human (CD63) (Santa Cruz, sc-5275) and rabbit polyclonal anti-human CD9 antibodies (abcam, ab223052), and at the same time, rabbit polyclonal anti-human HSP70 antibodies (ABclonal, A0284) was used as positive marker protein, and rabbit polyclonal anti-human Hsp90B1 antibodies (ABclonal, A0989) and rabbit polyclonal anti-human Calnexin antibodies (ABclonal, A4846) were used as negative marker protein. Mouse monoclonal anti-human GAPDH (abcam, ab8245) as a loading control. Purified exosome particles were washed with PBS and lysed with RIPA lysis buffer. The subsequent Western blotting procedure was consistent with the procedure of our previous study.15
Exosomal Protein Sample Preparation
The serum samples were extracted with SystemBiosciences (SBI) exosomes extraction kit (EXOTC50A-1), and the extracted exosomes samples were extracted with SDT (4% (w/v) SDS, 100mM Tris/HCl pH 7.6, 0.1M DTT) lysis method to extract protein and BCA method for protein quantification. Take an appropriate amount of protein from each sample and use the Filter aided proteome preparation (FASP) method for trypsin digestion, and then use the C18 Cartridge to desalt the digested peptides.
LC-PRM/MS Detection and Analysis
According to the pre-experimental results, the target peptides were subjected to PRM quantitative analysis. Import peptide information suitable for PRM analysis into the software Xcalibur for PRM method settings. Take 2ug peptides from each sample and mix with 20 fmol standard peptide, a total of 40 samples. Chromatographic separation was performed using the Easy nLC system with nanoliter flow rate HPLC. Buffer: A solution is a 0.1% formic acid aqueous solution, and B is a 0.1% formic acid acetonitrile solution (84% for acetonitrile). The column was balanced with 95% A liquid. After the sample was injected into the Trap Column, it passed through the Thermo scientific EASY column for gradient separation at a flow rate of 250 nl/min. The liquid phase separation gradient was as follows: 0 minutes-42 minutes, the linear gradient of liquid B was from 5% to 23%; 42 minutes-50 minutes, the linear gradient of liquid B was from 23% to 40%; 52 minutes-In 60 minutes, the linear gradient of liquid B rose to 100% and maintained. The samples separated by nano-upgraded high performance liquid chromatography were analyzed by Q-Exactive mass spectrometer (Thermo Scientific) for PRM mass spectrometry. Analysis time: 60 min, detection mode: positive ion. The scanning range of the primary mass spectrometer: 300–1800 m/z, mass spectral resolution: 60,000 (m/z 200), AGC target: 3e6, Maximum IT: 200 ms. After each full MS scan (full MS scan), 20 PRM scans (MS2 scans) were collected according to the Inclusion list, isolation window: 1.6 Th, mass spectrum resolution: 30,000 (m/z 200), AGC target: 3e6, Maximum IT: 120 ms, MS2 Activation Type: HCD, Normalized collision energy: 27. 40 samples were tested for PRM, and the original PRM files were analyzed by skyline 3.5.0.
Data Difference Analysis and Bioinformatics Analysis
Screen differentially expressed proteins (DEPs) with a fold change greater than 1.5 times (up-regulation greater than 1.5-fold or down-regulation less than 0.667-fold) and p-value less than 0.05. Perform GO (Gene Ontology) function annotation and enrichment analysis (including biological process, molecular function, cell component analysis) and cluster analysis of differential proteins. Based on the information in the STRING (http://string-db.org/) database, find direct and indirect interaction relationships between target proteins, generate interaction networks, and analyze the networks.
Statistical Analysis
All statistical analysis was performed using SPSS 22.0, and Graph Pad Prism 7 was used for image rendering. Student t test was used for analyzing the data conforming to normal distribution, while non-parametric test was used for analyzing the data not conforming to normal distribution. In all cases, P<0.05 was considered significant and P<0.01 was considered highly significant.
Results
Purification and Validation of Serum Exosomes from Human Peripheral Blood
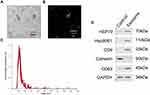
Exosomes purified from CHB, LC, and HCC patients serum were identified by transmission electron microscopy, ZetaView, and Western blotting, revealing a population of nanovesicles about 100 nm in size with typical morphology of exosomes (Figure 1A–C). The morphology of exosomes was complete and numerous, with a particle concentration of five million per milliliter of PBS (Figure 1C). In addition, Western blotting analysis of protein extracts from the isolated exosomes confirmed that these particles expressed the characteristic exosome-positive marker proteins CD9, CD63 and HSP70, and the negative markers Hsp90B1 and Calnexin expressed little or no expression (Figure 1D). The above results showed that exosomes were obtained with high purity and concentration.
Changes of Differentially Expressed Proteins (DEPs) in Serum Exosomes of Patients with CHB, LC and HCC
Through iTRAQ and Label-free quantitative proteomics in the early stage of our experiment, 11 DEPs were obtained for the next step of quantitative detection of PRM proteins. The PRM method can detect the unique peptide segment of the protein, and the change of the protein expression can be reflected by the change of the peptide segment (Figure 2). Then, we grouped the DEPs for statistical comparison (Table 3) to compare the expression differences of each protein in HCC patients before (AB group) and after (AA group) surgery and at different stages such as CHB and LC.
 |
Table 3 Differential Expression of Peptides in Serum Exosomes of Patients with CHB, LC and HCC Before (AB Group) and After Surgery (AA Group) |
Statistical analysis of DEPs according to different groups showed that compared with the CHB group, the significantly up-regulated protein in the preoperative liver cancer group was VWF (Table 3). Compared with the LC group, the significantly up-regulated proteins in the AB group were SELENOP, SERPIND1, and Hp (Table 3). Compared with the CHB group, the significantly up-regulated proteins in the AA group were CRP, FGA, and VWF (Table 3). Compared with the liver cirrhosis group, the significantly up-regulated proteins expressed in the hepatocellular carcinoma group were CRP and SHBG (Table 3). Compared with the CHB group, the significantly up-regulated protein in the LC group was VWF (Table 3). Compared with the CHB group, the significantly down-regulated proteins in the AB group were TTR and Hp (Table 3). Compared with the LC group, the significantly down-regulated protein in the AB group was APOA4 (Table 3). Compared with the CHB group, the significantly down-regulated proteins in the AA group were SERPINC1, APOA4, and TTR (Table 3). Compared with the LC group, the significantly down-regulated proteins in the AA group were SERPINC1 and APOA4 (Table 3). Compared with the CHB group, the significantly down-regulated protein in the LC group was Hp (Table 3).
In order to understand different proteins as postoperative predictors of HCC patients, 11 DEPs were grouped and counted according to the differences before and after surgery (Table 3). The results showed that compared with the AA group, the significantly up-regulated protein in the AB group was SERPIND1, and the significantly down-regulated proteins were CRP and SERPINA3 (Table 3). These DEPs have important implications for disease progression and prognosis in patients undergoing liver cancer surgery.
At the same time, according to the quantitative results obtained by PRM, we selected APOA4, HP and SERPIND1 to verify the protein expression by Western blotting in parallel. The results showed that the expression of APOA4 was down-regulated in AA group compared with the CHB and LC group, which was consistent with the PRM results (Figure 3). Compared with the CHB groups, the expression of Hp was down-regulated in the AB and LC groups, which was consistent with the PRM results. Compared with the LC and AA groups, the expression of SERPIND1 was up-regulated in the AB group, which was consistent with the PRM results. The results of Western blotting and PRM indicated that the protein expression results were reliable, which could relatively truly reflect the expression of DEPs in serum exosomes of liver diseases, and could provide a theoretical basis for clinical diagnosis and detection indicators.
 |
Figure 3 The relative expression of three DEPs (APOA4, HP and SERPIND1) in serum exosomes of patients with CHB, LC and HCC before (AB) and after (AA) surgical operation by Western blotting. |
Bioinformatics Analysis of DEPs in Serum Exosomes of Patients with CHB, LC and HCC
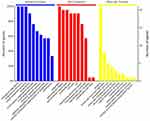
When conducting proteomics research, our research object is the collection of all proteins in cells, tissues or organisms. For subsequent functional research and analysis, it is important to understand the function of each protein, cell location, and the biological processes involved, but for high-throughput omics, understanding which functions or biological pathways are significantly affected by biology is the first task. Therefore, it is necessary to summarize and analyze the studied proteins and their functions from a more systematic and general perspective. GO enrichment analysis on the DEPs of each comparison group were performed by using Fisher’s exact test method. The results were shown in the Figure 4, this figure showed the top ten items of biological process (BP), cellular component (CC) and molecular function (MF) enrichment analysis significance ranking. The items of each category were sorted from left to right according to their -log(p-value) value, the more to the left, the more significantly. Among them, important biological processes such as biological regulation, response to stimulus, single organism process, localization, and multicellular organismal process, as well as positioning proteins such as extracellular region, extracellular region part, organelle part, cell part and molecular functions such as binding, molecular function regulator, transporter activity and structural molecular activity were significantly changed.
Cluster analysis is an exploratory data analysis method whose purpose is to group and classify data on the basis of similarity. In the results of clustering grouping, the data pattern similarity within the general group is relatively high, while the data pattern similarity between the groups is relatively low, so the grouping can be effectively distinguished. In this article, we used the pheatmap based on R language to analyze the heat map. In order to eliminate the difference in material values as much as possible when drawing the heat map, we used the Z-score algorithm to normalize the data. The more bluish the color in the heat map, the lower the content of the protein in the sample, on the contrary, the more reddish the color, the higher the content. The protein content of DEPs in each patient was showed in Figure 5.
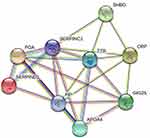
The proteins in the organism perform their functions by regulating and mediating each other. The study of the network formed by the interaction between proteins is of great significance for revealing the function of proteins. Among them, the highly aggregated proteins may have the same or similar functions; the highly connected proteins may be the key points affecting the metabolism or signal transduction pathways of the whole system. In the protein interaction network, nodes represent proteins, and lines represent interactions between proteins. The number of proteins that directly interact with a protein is called the degree of connectivity of the protein. Generally speaking, the greater the degree of connectivity of a protein, the greater the disturbance to the entire system when the protein changes. In this study, the DEPs of each comparison group were constructed to construct a protein interaction network diagram, and the results were shown in Figure 6. The result showed that Hp, TTR, SERPIND1, FGA and SERPINC1 were the core proteins of the network and interacted strongly with each other. These proteins may be the key to maintain the balance and stability of the system, and are the candidate proteins for subsequent key research.
Discussion
Diagnosis of liver disease is challenging, especially cirrhosis and HCC. HCC can be detected by ultrasound, nuclear magnetic resonance, CT imaging or serum AFP, and AFP is currently the best serum marker for diagnosing HCC. However, in some HCC patients with tumors less than 2 cm in diameter, AFP levels did not exceed the upper limit of detection, resulting in lower diagnostic accuracy. Among the 25 HCC patients we recruited, 20 had tumor diameter less than 2 cm, only 3 had AFP detection level greater than 100 ng/mL, and the early development of HCC had no obvious symptoms, making it difficult to diagnose accurately. Therefore, early diagnosis of HCC can greatly improve the survival rate of patients.
Liquid biopsy is the most important diagnostic and therapeutic technique that can be used in cancer research and dynamically reflect the full picture of tumor gene spectrum in clinical practice. Exosomes are spherical vesicles secreted by cells, rich in lipids, proteins, RNA and DNA,1,2 which mediate information transfer between cells by transferring different biologically active molecules. There is good evidence that exosomes are associated with a variety of liver diseases.3–8 In addition, almost all types of cells in the body can secrete exosomes. Therefore, microRNAs (miRNAs) and proteins in exosomes will become important biomarkers for the diagnosis and treatment of liver diseases such as HCC. We screened serum samples from 22 patients with CHB, 10 patients with LC and 22 patients with HCC from the sample library, isolated and purified exosomes, and identified exosomes by Zetaview particle size analysis, Western blotting and transmission electron microscopy. The morphological structure and marker proteins of exosomes proved that the isolated microvesicles were exosomes.
In the early stage, we screened the DEPs from the serum exosomes of patients with CHB and HCC by two methods of Label-free and iTRAQ, verified the expression of the DEPs by the PRM method, and classified the 11 kinds of DEPs according to different groups. The results showed that the DEPs screened by PRM mass spectrometry were closely related to the development of liver disease. It is well known that Hp is an acute phase protein. It is mainly produced by the liver and secreted into the blood. Studies have shown that Hp plays an important role in hemoglobin metabolism and may be involved in inflammation and host defense responses to infection.16 Recent studies have found that Hp plays a role in angiogenesis.16 A large number of studies have shown that Hp is expressed in malignant ovarian epithelial cells, renal tumor cells and HCC tissues,16 suggesting that serum Hp may be a biomarker for cancer diagnosis. However, the concentration of Hp in the serum of HCC patients is currently unknown. The results of PRM in this study showed that compared with the LC group, Hp in serum exosomes of HCC patients was significantly up-regulated, and the results of Western blotting were consistent with the PRM results. In addition, Ang et al also conducted a study of elevated serum Hp concentrations in HCC patients,16 which is consistent with our results and strongly suggests that serum Hp is a potential diagnostic biomarker for HCC. TTR is an extracellular protein mainly produced in the liver and choroid plexus and has a stabilizing role in the transport of thyroxine and retinol throughout the body and the brain. Studies have shown that TTR plays a role in tumor progression mainly by regulating the proliferation of endothelial cells and immune cells surrounding the tumor. Therefore, TTR has recently been described as a biomarker for lung and ovarian cancers, as its levels were shown to be elevated in the serum of patients.17 TTR has been widely used as a biomarker for malnutrition, health conditions in the elderly, and diseases including metabolic, inflammatory, and septic because liver gene expression is shown to decrease in these conditions.17 Our results showed that TTR was significantly down-regulated in serum exosomes of HCC patients compared with CHB patients, which was consistent with the trend of the above results, suggesting that TTR in serum exosomes has the potential to become a biomarker.
In order to understand different proteins as postoperative predictors of HCC patients, we screened out the proteins CRP, SERPINA3 and SERPIND1 that were significantly differentially expressed in HCC patients before and after surgery. Studies have shown that serum CRP is a prognostic factor for overall survival (OS) and recurrence of HCC in patients undergoing resection or non-surgical treatment.18 Furthermore, studies have shown that elevated CRP in serum is associated with HCC recurrence after liver transplantation and may be a marker of tumor biology. Studies have shown that CRP plays an important role in the occurrence, development and/or prognosis of various cancers such as esophageal squamous cell carcinoma, cervical cancer, and non-small cell lung cancer.18 In HCC, CRP predicts poor overall survival and recurrence after liver resection and is associated with poor prognosis after living donor liver transplantation.18 The results in this study showed that compared with the patients before liver cancer surgery, the expression of CRP in the serum exosomes of the patients after surgery was significantly up-regulated, which also confirms the above research results, suggesting that the CRP in the serum exosomes of HCC has the potential to be a prognostic indicator in patients with liver cancer surgery. Another protein is SERPINA3, a member of the serine protease inhibitor gene family that has been shown to be involved in anti-inflammatory therapy and is associated with many human diseases.19 Studies have shown that SERPINA3 mRNA levels are lower in HCC tumors compared to non-tumors.20 In this study, SERPINA3 was significantly up-regulated in serum exosomes of patients after liver cancer surgery compared with patients before liver cancer surgery, suggesting that SERPINA3 can be used as a prognostic indicator for patients with liver cancer surgery to some extent. Heparin cofactor 2 (SERPIND1) is another heparin-dependent antithrombin protein other than antithrombin III in plasma, and is a new component of the physiological anticoagulation system. SERPIND1 belongs to the serine protease inhibitor family. In recent years, its role in cancer has gradually attracted the interest of researchers. The study found that SERPIND1 expression was significantly elevated in epithelial ovarian cancer, suggesting that SERPIND1 may be an effective marker for evaluating the prognosis of ovarian cancer.21 However, the role of SERPIND1 in HCC development remains poorly understood. We found that SERPIND1 was significantly up-regulated in serum exosomes of HCC patients before surgery compared with LC patients and patients after HCC surgery. This study is the first to study the expression of SERPIND1 in serum exosomes of HCC, and its expression was significantly higher in patients with HCC before and after surgery. The difference can be further investigated for its possibility as a prognostic indicator for HCC surgery. Based on the results of this study, we need to further expand the number of enrolled samples to further verify the results. More research is needed in the future to realize the transformation from basic research to clinical application.
Conclusion
The DEPs were validated according to the PRM technique and Western blotting, and the results showed that Hp and TTR were expected to be the marker proteins for early diagnosis of HCC. CRP, SERPINA3 and SERPIND1 have the potential to be prognostic indicators for liver cancer surgery. We need to further expand the number of enrolled samples to further verify the results. More research is needed in the future to realize the transformation from basic research to clinical application.
Data Sharing Statement
Technical appendix, statistical code, and dataset were available from the corresponding author at [email protected] and [email protected]. Participants gave informed consent for data sharing.
Ethics Approval and Informed Consent
The samples in this study were obtained with informed consent. The study was approved by Ethics Committee of the Dalian Public Health Clinical Center (Approval ID: 2020-032-001). The study complies with the Declaration of Helsinki.
Acknowledgments
We are grateful for the biological samples provided by the Department of Biobank of Dalian Public Health Clinical Center, Dalian, China.
Funding
This work was supported by the Dalian Science and Technology Innovation Fund (No. 2021JJ13SN71); Dalian Medical Science Research Program (No. 2011015); and the Natural Science Foundation of Liaoning Province (No. 2019-ZD-1003).
Disclosure
The authors declare the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Kalluri R. The biology and function of exosomes in cancer. J Clin Investig. 2016;126(4): 1208–1215. doi:10.1172/JCI81135
2. Morelli AE, Larregina AT, Shufesky WJ, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266.
3. Longatti A, Boyd B, Chisari FV. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J Virol. 2015;89:2956–2961.
4. Yang Y, Han Q, Hou Z, Zhang C, Tian Z, Zhang J. Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell Mol Immunol. 2017;14:465–475.
5. Wei JX, Lv LH, Wan YL, et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284–1294.
6. Huang G, Brigstock DR. Regulation of hepatic stellate cells by connective tissue growth factor. Front Biosci. 2012;17:2495–2507.
7. Witek RP, Yang L, Liu R, et al. Liver cell-derived microparticles activate hedgehog signaling and alter gene expression in hepatic endothelial cells. Gastroenterology. 2009;136:320–30 e2.
8. Koeck ES, Iordanskaia T, Sevilla S, et al. Adipocyte exosomes induce transforming growth factor beta pathway dysregulation in hepatocytes: a novel paradigm for obesity-related liver disease. J Surg Res. 2014;192:268–275.
9. Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887.
10. Pisitkun T, Shen RF, Knepper MA Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004; 101(36):13368–13373. doi:10.1073/pnas.0403453101
11. Ogawa Y, Miura Y, Harazono A, et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull. 2011;34:13–23.
12. Masyuk AI, Huang BQ, Ward CJ, et al. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol. 2010;299:G990–9.
13. Lässer C, Alikhani VS, Ekström K, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9.
14. Asea A, Jean-Pierre C, Kaur P, et al. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J Reprod Immunol. 2008;79:12–17.
15. Pu C, Huang H, Wang Z, et al. Extracellular vesicle-associated mir-21 and mir-144 are markedly elevated in serum of patients with hepatocellular carcinoma. Front Physiol. 2018;9:930.
16. Ang IL, Poon TC, Lai PB, et al. Study of serum haptoglobin and its glycoforms in the diagnosis of hepatocellular carcinoma: a glycoproteomic approach. J Proteome Res. 2006;5:2691–2700.
17. Magalhães J, Eira J, Liz MA. The role of transthyretin in cell biology: impact on human pathophysiology. Cell Mol Life Sci. 2021;78:6105–6117.
18. Meischl T, Rasoul-Rockenschaub S, Györi G, et al. C-reactive protein is an independent predictor for hepatocellular carcinoma recurrence after liver transplantation. PLoS One. 2019;14:e0216677.
19. Zhang Y, He J, Zhao J, et al. Effect of ApoA4 on SERPINA3 mediated by nuclear receptors NR4A1 and NR1D1 in hepatocytes. Biochem Biophys Res Commun. 2017;487:327–332.
20. Ko E, Kim JS, Bae JW, Kim J, Park SG, Jung G. SERPINA3 is a key modulator of HNRNP-K transcriptional activity against oxidative stress in HCC. Redox Biol. 2019;24:101217.
21. Guo Q, Zhu L, Wang C, et al. SERPIND1 affects the malignant biological behavior of epithelial ovarian cancer via the PI3K/AKT pathway: a mechanistic study. Front Oncol. 2019;9:954.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.