Back to Journals » Risk Management and Healthcare Policy » Volume 16
Identification of the Genotypes Circulating in the Ecuadorian Population Infected with the Hepatitis C Virus (HCV)
Authors Moncayo M, Teran E , Reyes J , Yerovi G, Robalino M, Aguilar AC, Garzon-Chavez D
Received 4 April 2023
Accepted for publication 8 July 2023
Published 3 August 2023 Volume 2023:16 Pages 1403—1409
DOI https://doi.org/10.2147/RMHP.S412599
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Jongwha Chang
Miguel Moncayo,1,2 Enrique Teran,1,3 Jorge Reyes,4,5 Gabriela Yerovi,6 Marcia Robalino,6 Ana Cristina Aguilar,1,3 Daniel Garzon-Chavez1,3
1Instituto de Microbiología, Universidad San Francisco de Quito USFQ, Quito, Ecuador; 2Laboratorio Clinico Pasteur Dr. Alberto Moncayo Calero, Quito, Ecuador; 3Colegio de Ciencias de la Salud, Universidad San Francisco de Quito USFQ, Quito, Ecuador; 4Facultad de Ciencias Químicas, Universidad Central del Ecuador, Quito, Ecuador; 5Departamento de Microbiologı´a, Hospital del IESS Quito Sur, Quito, Ecuador; 6Ministerio de Salud Pública del Ecuador, Quito, Ecuador
Correspondence: Daniel Garzon-Chavez, Email [email protected]
Introduction: The hepatitis C virus (HCV) is responsible for 1.5 million new infections, and around 290 thousand deaths worldwide. 15 to 30% of the patients that go into a chronic phase of the disease will develop cirrhosis or hepatocellular carcinoma within 20 years and is the leading etiology for liver transplantation. HCV genetic characteristics display a remarkable genetic diversity, which divides HCV into 8 genotypes and 67 subgenotypes; the treatment and probability of chronic HCV depend on these genotypes and subgenotypes. In Ecuador, there is no available information regarding HCV genotypes and subgenotypes; therefore, this study aims to provide an overview of the main genotypes circulating in Ecuador.
Methods: In a cross-sectional and descriptive study using the Ecuadorian Ministry of Health (MSP) registry of patients already diagnosed with Hepatitis C (HCV) between 2017 and 2019. From 51 patients identified by health ministry, blood samples from a total of 15 subjects (named HCV1 to HCV15) were collected using an appropriate venipuncture technique. Pandemic-related circumstances avoid reaching all patients identified by health ministry.
Results: After the amplification of 11 samples from patients living in the Ecuadorian territory, the genotypes of HCV obtained were distributed as follows: 6 samples corresponding to subgenotype 2b (54.5%), 2 samples corresponding to subgenotype 1a (18.2%), 2 samples corresponding to subgenotype 4d (18.2%) and 1 corresponding to sample 1b (9.1%).
Conclusion: These results represent the first epidemiological approach to genotype distribution in Ecuador, and it contributes to better management of patients. We emphasize the importance of the development of better strategies from the Healthcare Ministry of Ecuador (MSP) for the identification, treatment and tracking of HCV patients.
Keywords: hepatitis C virus, hepatitis C genotype, hepatitis C subgenotype, hepatitis C prevalence, cirrhosis, hepatocellular carcinoma
Introduction
Hepatitis C virus (HCV) is a single-stranded, positive-sense RNA virus of approximately 9600 nucleotides in length.1 The NS5B gene is an RNA-dependent RNA polymerase located between the 7597 and 9413 base pairs; this gene lacks proofreading ability. Therefore, HCV represents a remarkable genetic diversity that divides HCV into 8 genotypes and 67 subgenotypes.2,3
Between 15% and 45% of patients infected with HCV can clear it in about 6 months without receiving any treatment.4 The remaining patients might develop, within 20 years, a chronic phase compromising cirrhosis or hepatocellular carcinoma.4,5 According to the World Health Organization (WHO), HCV is responsible for approximately 1.5 million new infections per year, and around 290 thousand deaths in 2019.4 HCV represents a significant public health problem, and in 2016 WHO declared it a global target, with different strategies to control HCV have been developed, highlighting a reduction of 90% in new cases of viral hepatitis, a reduction of 60% in deaths related hepatitis by 2030, 80% of eligible patients with chronic HCV with ongoing treatment.6
In addition, it is crucial to identify the genotyping in HCV because treatment depends on genotype and subgenotype, and it can also influence the treatment duration, and clearance rate.7 However, in Ecuador currently, there are no data about HCV circulation genotypes but in neighboring countries like Colombia and Peru genotype 1 is prevalent (88.5% and 86%, respectively),5 while in others like Venezuela genotype 2 represents 34.4% of the cases.5 Therefore, this study aimed to provide a first approximation of the main genotypes circulating in Ecuador.
Methods
In a cross-sectional and descriptive study using the Ecuadorian Ministry of Health (MSP) registry of patients already diagnosed with Hepatitis C (HCV) between 2017 and 2019, diagnose was previously done by MSP health services, and patients were included under the following inclusion criteria: 1) confirmed diagnosis of chronic HCV infection with persistent viral charge after 6 months, all patients included in the present study were diagnosed by specific viral charge test according to the American Association for the Study of Liver Diseases Hepatitis C Guidance 2019 Update,8 2) age over 18 years old, and 3) written informed consent. The exclusion criteria were 1) previous HCV diagnosis but not being part of the MSP database from year 2017 to 2019, 2) patients under the age of 18 years, 3) patients that decide not to provide written informed consent. From 51 patients identified by health ministry, blood samples from a total of 15 subjects (named HCV1 to HCV15) were collected using an appropriate venipuncture technique. Pandemic-related circumstances avoid reaching all patients identified by health ministry. Samples were later centrifuged, the plasma was placed in Eppendorf tubes and transported in a nitrogen tank, and later stored at −80 °C at the School of Medicine in the Universidad San Francisco de Quito.
Ethical Statement
The study was approved by the Human Research Ethics Committee at the Universidad San Francisco de Quito (2018-241IN) and by the Ministry of Public Health of Ecuador. We confirm that all patients provided informed consent and the study was conducted in accordance with the Declaration of Helsinki.
Primers Selection
Primers for the amplification of the NS5B section of the HCV genome were selected from the work of Tong et al 2015.9 We used primers named F1, R1 to F2, R2 of the NS5B protein (Figure 1).
 |
Figure 1 (a) Graphic made using Genious Prime 2022.1.1, location of primers used on HCV NS5B protein region (b) primers used for NS5B gene amplification as described by Tong YQ, Liu B, Liu H, Zheng HY, Gu J, Liu H, et al. Accurate genotyping of hepatitis C virus through nucleotide sequencing and identification of new HCV subtypes in China population. Clin Microbiol Infect. 2015;21:874.e9–874.e21.9 |
HCV RNA Extraction
The RNA extraction of the samples was performed using MagMAXTM Viral/Pathogen Nucleic Acid Isolation Kit following the manufacturer’s instructions (ThermoFisher Scientific Inc).
cDNA Synthesis
The complementary DNA (cDNA) was synthesized with the Thermo Scientific RevertAID First Strand cDNA Synthesis Kit, using the HCV RNA previously obtained following the manufacturer’s instructions (ThermoFisher Scientific Inc).
HCV NS5B Gene Fragment Amplification by PCR
The HCV cDNA template was used for the amplification of the NS5N gene fragment by PCR as Figure 1. The PCR was performed using InvitrogenTM PlatinumTM II Hot-Start Green PCR Master Mix following the manufacturer’s instructions (ThermoFisher Scientific Inc). The PCR conditions used were the following: 94°C for 3 min; 94°C for 30 sec, 56°C for 40 seg, 72°C for 60 sec, 35 cycles; 72°C for 10 min. The temperature of annealing of 56°C was selected as described by Tong et al. Then, the amplicons were quantified using the EPOCH Microplate Spectrophotometer Reader. After quantification, the amplicons were sent to Macrogen, South Korea, for Sanger sequencing. Nucleotide sequences obtain were entered into GenBank under accession numbers: ON540736, ON540737, ON540738, ON540739, ON540740, ON540741 ON540742, ON540743, ON540744, ON540745 and ON540746.
Phylogenetic Analysis
Sequences reported were compared to identify local similarity regions between sequences using BLAST. Phylogenetic trees were built using the Neighbor-Joining method and Tamura-Nei genetic distance model using the software Geneious Prime 2022.0.1. Reference sequences added to the phylogenetic tree were obtained by searching in the NCBI GenBank using the keywords “HCV” AND “NS5B” OR “complete genome”.
Results
From the 51 patients with HCV reported by the MSP data during 2017 to 2019, 15 patients met the inclusion criteria and attended to MSP recruitment; 9 females (60%) and 6 males (40%); mean age 52 ± 24.5. Patient ethnic group self-identification was 100% mestizo. Geographical distribution of samples was Quito 60% and Guayaquil 40% (Table 1). Only 11 of the 15 samples amplified NS5B section. The genotype and subgenotype of 8 samples (HCV1 HCV3, HCV4, HCV7, HCV10, HCV11, HCV13, HCV14), were determined using primers F1-R1 (forward and reverse; Figure 2). Then, using the F2-R2 forward and reverse fragments of the sequences was possible to determine that one subject (HCV9) belonged to the subgenotype 2b (Supplementary Figure 1), this sample did not amplify with primers F1-R1.
 |
Figure 2 Phylogenetic tree based on fragments F1-R1 forward and reverse, determination of genotypes and subgenotypes. |
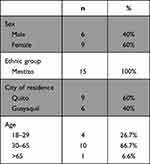 |
Table 1 Sociodemographic Characteristic of HCV Ecuadorian Patients |
From sequences HCV2 and HCV15 only was possible to amplify a single strand of the genome sections, F1-R1 (forward strand) for subject HCV2 and F1-R1 (reverse strand) for the subject HCV15. Reference sequences were obtained from the NCBI GenBank with the keywords previously described. To compare reference sequences with HCV2 and HCV15 and provided an accurate phylogenetic analysis, the reference sequences were aligned and trimmed. The section used of the reference sequences for phylogenetic analysis of the sample HCV2 included bases from 8135 to 8357, while for HCV15 the section used of the reference sequences for phylogenetic analysis included bases 8144 to 8408. Later, obtained sections were used to create individual phylogenetic trees and the results showed that subject HCV2 belonged to the subgenotype 2b (Supplementary Figure 2) and the subject HCV15 did to subgenotype 1a (Supplementary Figure 3), the results obtained in the phylogenetic analysis for HVC2 and HCV15 were in agreement with the results from BLAST.
Samples of six subjects corresponded to subgenotype 2b (54.5%), two samples to subgenotype 1a (18.2%), two samples to subgenotype 4d (18.2%), and one sample belonged to subgenotype 1b (9.1%).
Discussion
This report shows, for the first time, that HCV subgenotype 2b is prevalent in our study sample (54.5%). This finding contradicts the data from Latin America compiled by Petruzziello et al,5 where genotype 1 (74.3%) is prevalent. In Ecuador’s neighboring countries, genotype 1 is also the most prevalent in Colombia (88.6%)10 and Peru (86%).11 Genotype 2 highest prevalence was found in Venezuela (34.4%), Argentina (24.7%), and Mexico (21.8%)5 but none as high we found.
Although in Latin America, it is estimated that 0.5% of its total population (ie, 3.8 million people) are infected with HCV,6 in Ecuador, according to the Ministry of Health from 2017 to 2019 there were 51 new cases (MSP, 2019). Therefore, the official prevalence of HCV in Ecuador period 2017–2019 according to the health authority was 0.0003%. This data is far below the reported in Colombia (0.9–1.1%)12,13 and Peru (0.1–0.5%),12–14 the two neighboring countries of Ecuador. Also, compared with data from other countries, where prevalence ranges from 0.3 to 3.3%,11 this data needs further studies to elucidate the real situation of HCV in Ecuador. It can be of great value to study and relate the information from blood banks regarding the anti-hepatitis C antibodies in asymptomatic population and the Ministry of Health’s database. Based on the study carried out by Grijalva et al,15 it is possible to seek to improve the safety of blood donation and the collection of data related to pathologies in Ecuadorian territory, thus obtaining a better understanding of HCV reality in Ecuador.
On the other hand, response to treatment, particularly to interferon (INF), is different between genotypes 1 and 2.16,17 Particularly, 50% of subjects with subgenotype 2a shows pharmacological susceptibility to INF, while those subjects with subgenotype 1b respond only in 11.1% of the cases.16,17 However, the combination of ribavirin with INF leads to a sustained virologic response, mainly in genotypes 2 and 3 (76–82%) than in genotypes 1, 4, 5, or 7 (42–52%).16,17
Development of multiple direct-acting antivirals (DAAs) which are defined by their mechanism of action and therapeutic target, as nonstructural proteins 3/4A (NS3/4A) protease inhibitors (PIs), NS5B nucleoside polymerase inhibitors (NPIs), NS5B non-nucleoside polymerase inhibitors (NNPIs), and NS5A inhibitors have revolutionized HCV infected patients’ treatment.17 However, in most developing countries, like Ecuador, the high cost of DAAs limited their use, and the lack of accurate epidemiologic information interferes with proper monitoring and treatment of patients.
The main limitation of our report was the small number of samples, however, taking into consideration the reported prevalence, we were able to get a huge proportion of the infected subject. It is important to emphasize that this initial study, with enough methodological rigor, raises the need to improve active diagnostic strategies and at the same time, the urgent need for DAAs availability to improve the quality of life for the patients.
Data Sharing Statement
Datasets that were analyzed or used in this study are available as Supplementary Information Supplementary Table 1, also all the sequences are available at the NCBI GenBank.
Author Contributions
All authors made significant contributions to the reported work, whether it was in the conception, study design, execution, data acquisition, analysis and interpretation, or in all of these areas. They participated in drafting, revising, or critically reviewing the article. They also gave their final approval of the version to be published, agreed on the journal to which the article has been submitted, and agreed to be accountable for all aspects of the work.
Funding
Financial support for this research was provided by Universidad San Francisco de Quito USFQ through a Med School Grants program - HUBI 15750.
Disclosure
Authors declare that they have no competing interests.
References
1. Tellinghuisen TL, Rice CM. The molecular virology of hepatitis C virus. Princ Med Biol. 2004;15(C):455–495.
2. Qiu P, Cai XY, Wang L, Greene JR, Malcolm B. Hepatitis C virus whole genome position weight matrix and robust primer design. BMC Microbiol. 2002;2:1–7. doi:10.1186/1471-2180-2-29
3. Tsukiyama-Kohara K, Kohara M. Hepatitis C virus: viral quasispecies and genotypes. Int J Mol Sci. 2018;19(1):1–8.
4. World Health Organization. Hepatitis C. World Health Organization; 2021. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c.
5. Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: an update of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22(34):7824–7840. doi:10.3748/wjg.v22.i34.7824
6. Blach S, Zeuzem S, Manns M, et al. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modeling study. Lancet Gastroenterol Hepatol. 2017;2(3):161–176. doi:10.1016/S2468-1253(16)30181-9
7. Messina JP, Humphreys I, Flaxman A, et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61(1):77–87. doi:10.1002/hep.27259
8. Ghany MG, Morgan TR; AASLD‐IDSA Hepatitis C Guidance Panel. Hepatitis C guidance 2019 update: American Association for the study of liver diseases-Infectious Diseases Society of America Recommendations for testing, managing, and treating hepatitis C virus infection. Hepatology. 2020;71(2):686–721. doi:10.1002/hep.31060
9. Tong YQ, Liu B, Liu H, et al. Accurate genotyping of hepatitis C virus through nucleotide sequencing and identification of new HCV subtypes in China population. Clin Microbiol Infect. 2015;21(9):874.e9–874.e21. doi:10.1016/j.cmi.2015.05.034
10. Santos Ó, Gómez A, Vizcaíno V, et al. Genotipos circulantes del virus de la hepatitis C en Colombia. Biomedica. 2017;37(1):22–27. doi:10.7705/biomedica.v37i1.3173
11. Sanchez JL, Abdel-Hamid M, Callahan JD, et al. Hepatitis C in Peru: risk factors for infection, potential iatrogenic transmission, and genotype distribution. Am J Trop Med Hyg. 2000;63(5):242–248. doi:10.4269/ajtmh.2000.63.242
12. Imbeth-Acosta P, Leal-Martínez V, Ramos-Clason E, et al. Prevalence of chronic infection by hepatitis C virus in asymptomatic population with risk factors in Cartagena, Colombia. Front Med. 2022;9:1–6. doi:10.3389/fmed.2022.814622
13. Maaroufi A, Vince A, Himatt SM, et al. Historical epidemiology of hepatitis C virus in select countries—volume 4. J Viral Hepat. 2017;24:8–24. doi:10.1111/jvh.12762
14. Farfán G, Cabezas C. Prevalence of viral hepatitis type C in blood donors in Peru. Revista de gastroenterología del Perú. 2003;23(3):171–176.
15. Grijalva MJ, Chiriboga RF, Vanhassel H, et al. Improving the safety of the blood supply in Ecuador through external performance evaluation of serological screening of blood donors. J Clin Virol. 2005;34(SUPPL. 2):S47–S52. doi:10.1016/S1386-6532(05)80034-4
16. Spengler U. Direct antiviral agents (DAAs) - A new age in the treatment of hepatitis C virus infection. Pharmacol Ther. 2018;183:118–126. doi:10.1016/j.pharmthera.2017.10.009
17. Jakobsen JC, Nielsen EE, Feinberg J, et al. Direct-acting antivirals for chronic hepatitis C. Cochrane Database Syst Rev. 2017;2017(6):154.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
