Back to Journals » International Journal of General Medicine » Volume 14
Identification of Prognostic Factors Related to Super Enhancer-Regulated ceRNA Network in Metastatic Lung Adenocarcinoma
Authors Li M , Yang B, Li X, Ren H, Zhang L, Li L, Li W, Wang X, Zhou H, Zhang W
Received 2 August 2021
Accepted for publication 16 September 2021
Published 1 October 2021 Volume 2021:14 Pages 6261—6275
DOI https://doi.org/10.2147/IJGM.S332317
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Mingjiang Li,1 Bo Yang,1 Xiaoping Li,1 Haixia Ren,2 Liang Zhang,1 Lei Li,1 Wei Li,1 Xuhui Wang,1 Honggang Zhou,3 Weidong Zhang1
1Department of Thoracic Surgery, Tianjin First Central Hospital, Tianjin, People’s Republic of China; 2Department of Pharmacy, Tianjin First Central Hospital, Tianjin, People’s Republic of China; 3College of Pharmacy, Nankai University, State Key Laboratory of Medicinal Chemical Biology, Tianjin, People’s Republic of China
Correspondence: Mingjiang Li; Weidong Zhang
Department of Thoracic Surgery, Tianjin First Central Hospital, Tianjin, 24 Fukang Road, Tianjin, 300192, People’s Republic of China
Tel +86-22-23627068
Email [email protected]; [email protected]
Introduction: The regulatory mechanisms of super enhancers (SEs) and ceRNA networks in LUAD progression are not well understood. We aimed to discover the prognostic-related ceRNA network regulated by SEs in metastatic LUAD.
Methods: RNA-seq data were extracted from The Cancer Genome Atlas (TCGA) database. Differentially expressed (DE) RNAs were identified by edgeR. CeRNA network was predicted and visualized using starBase and Cytoscape. H3K27ac ChIP-seq data were derived from the Gene Expression Omnibus (GEO) database, and used for SE identification. Kaplan–Meier curve and multivariate Cox model were applied for prognostic analysis. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein–protein interaction (PPI) network were performed for functional analysis. SEs of AC074117.1 were verified by ChIP-qPCR in A549 and H1299 cells. MTT assay was performed to analyze cell proliferation. Luciferase activity assay was carried out to validate the target targeting relationships of ceRNA network.
Results: A total of 2355 DEmRNA, 483 DElncRNA and 155 DEmiRNA were identified between metastatic LUAD and adjacent normal tissues. CeRNA network consisting of 7 DElncRNAs, 18 DEmiRNAs and 15 DEmRNAs was constructed. Among the seven DElncRNAs in ceRNA network, only AC074117.1 was regulated by SEs. SE-regulated prognostic ceRNA sub-network consisting of FKBP3, E2F2, AC074117.1 and hsa-let-7c-5p was screened and verified. The overlapping co-expressed mRNAs of FKBP3, E2F2, AC074117.1 and hsa-let-7c-5p were mainly related to cell division and Fanconi anemia pathway. Genes in the ceRNA sub-network were correlated with DNA mismatch repair markers. Functional experiments proved that AC074117.1 was highly expressed in LUAD cells. AC074117.1 silencing notably inhibited proliferation of A549 and H1299 cells. Luciferase activity assay confirmed the direct relationship in AC074117.1-hsa-let-7c-5p-FKBP3/E2F2 network.
Conclusion: A novel prognostic ceRNA sub-network regulated by SEs was identified in metastatic LUAD. This study provided potential therapeutic targets and prognostic markers for further study of metastatic LUAD.
Keywords: lung adenocarcinoma, super enhancer, prognosis, competitive endogenous RNA, differentially expressed genes
Introduction
Lung cancer is one of the most serious malignant tumors in the world.1 It mainly occurs in middle-aged and elderly populations. Moreover, the prevalence of young patients has shown an upward trend in recent years.2 Lung cancer can be divided into non-small cell lung cancer (accounts for 80–84% of lung cancer cases) and small cell lung cancer.1 Lung adenocarcinoma (LUAD) is the most prevalent pathological subtype of non-small cell lung cancer.3 Despite the advancement in targeted therapies and immunotherapy for lung cancer in recent years, the 5-year survival rate of LUAD remains less than 20%.4 Exploring the molecular mechanism underlying LUAD development is crucial for seeking novel biomarkers of LUAD.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNA with more than 200 nucleotides in length, accounting for 68% of non-coding RNA.5 Initially, lncRNAs had been considered as “junk RNA” in genome due to no or limited protein-coding capacity.6 However, lncRNAs have been found to play important roles in human diseases including cancer, diabetes and cardiovascular disease.7–9 LncRNAs are key participants in the regulation of gene expression, which functions as the competitive endogenous RNAs (ceRNAs) to sponge miRNAs, and reduce the inhibitory effects of miRNAs on their targets.10,11 CeRNA network as a whole rather than individual transcripts affects post-transcriptional regulation. The imbalance of ceRNA network may contribute to cancer development.12 There are a large number of dysregulated lncRNAs in LUAD. These abnormal lncRNAs regulate cancer progression through ceRNA mechanism.13–15
Super enhancers (SEs) are an enhancer cluster of 8–20 kb in length, enriched with a high-density of key transcription factors and cofactors, which are important regulatory elements in maintaining the characteristics of cancer cells.16 SEs are capable of promoting oncogene transcription and cancer progression by regulating non-coding RNA transcription.17 Screening of lncRNAs regulated by SEs is helpful to a comprehensive understanding of the regulatory mechanism of ceRNA networks. Exploration of the ceRNA networks regulated by SEs could provide new ideas to reveal the complex mechanism underlying the malignant process of LUAD.
In this study, differentially expressed (DE) RNAs between metastatic LUAD and adjacent normal tissues were screened based on TCGA database. A ceRNA network was constructed using the DERNAs. SEs and the lncRNAs regulated by SEs in ceRNA network were filtered. The prognostic ceRNA sub-network regulated by SEs was identified. The purpose of this study was to identify the prognostic factors associated with the SE-regulated ceRNA network in metastatic LUAD.
Methods
Data Collection
Transcriptome profiling containing mRNA, lncRNA and miRNA of 507 LUAD patients (including 67 metastatic LUAD patients) were downloaded from The Cancer Genome Atlas (TCGA) database. Relevant prognostic information of the 507 patients with LUAD was also collected from TCGA database.
Identification of DERNAs
DElncRNAs, DEmiRNAs and DEmRNAs between LUAD tumor and adjacent normal tissues were determined using edgeR package in R software. The cut-off values were set to |log fold change| (|logFC|)>1 and P<0.05.
Construction of ceRNA Network
All of the DElncRNAs, DEmiRNAs and DEmRNAs were used for ceRNA network construction. Interactions of miRNA with lncRNA or mRNA were predicted with starBase v2.0 (http://starbase.sysu.edu.cn/). CeRNA network was visualized using Cytoscape v3.7.2.
SE Analysis
H3K27ac ChIP-seq data of HSC4, A549 and H23 cells were obtained from GEO database (GSE143653). H3K27ac score was calculated using FindPeaks tool in Homer algorithm. Enhancer clusters were sorted according to the H3K27ac score using super enhancer tool in Homer algorithm. Enhancer clusters with a slope of tangent greater than 1 were considered to be SEs. Gene closest to SE on the genome was defined as SE-associated gene. The common SE-associated genes of HSC4 and A549 cells were screened using a Venn diagram. The classification of SE-associated genes was represented in a pie chart.
Survival Analysis of Genes in ceRNA Network
Patients with LUAD were divided in to high- and low-expression groups according to the quartile of gene expression levels. Kaplan–Meier method and the Log rank test were used to evaluate the effect of gene expression on overall survival, with P<0.05.
Construction of Prognostic ceRNA Sub-Network and Risk Score System
Prognostic ceRNA sub-network was constructed based on the prognostic genes in ceRNA network. GDCRNATools package and Cytoscape software were used for sub-network prediction and visualization.
Genes in the prognostic ceRNA sub-network were further used for multivariate Cox analysis. Risk score was calculated according to the formula: risk score = (β1*RNA1 expression level) + (β2*RNA2 expression level) + (β3*RNA3 expression level) + (β4*RNA4 expression level). Patients were stratified into high- and low-risk groups based on the median of risk score. Time-dependent receiver operating characteristic (ROC) curve and area under the curve (AUC) were obtained using ROC package in R software to assess prognostic accuracy and specificity.
Functional Analysis and Protein–Protein Interaction (PPI) Network Construction
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were performed using ClusterProfile package in R software. P<0.05 was considered to be significant. PPI was analyzed using STRING database v11.0 (http://www.string-db.org/), and then visualized using Cytoscape software.
Cell Culture and Transfection
One normal human bronchial epithelial cell line (BEAS-2B) and six LUAD cell lines (H460, HCC827, A549, H1299, PC9 and Calu3) were obtained from American Type Culture Collection (ATCC, VA, USA). All of these cells were cultured in DMEM medium (Invitrogen, CA, USA) at 37 °C in 5% CO2. ShRNAs (sh-control, sh-AC074117.1#1 and sh-AC074117.1#2) were synthesized by GenePharma (Shanghai, China). Sh-control, sh-AC074117.1#1 and sh-AC074117.1#2 were transfected into A549 or H1299 cells using Lipofectamine 2000 (Invitrogen). The sequences of sh-AC074117.1#1 and sh-AC074117.1#2 were as following: sh-AC074117.1#15ʹ-CACCGTTCAGTGATGAGTGGGTCCAGCGAACTGGACCCACTCATCACTGAA-3ʹ; sh-AC074117.1#25ʹ-CACCGAAGACCCGTATAAAGAACCAACGAATTGGTTCTTTATACGGGTC −3ʹ.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA of A549 and H1299 cells were isolated by Trizol (Invitrogen). Reverse transcription was carried out by Prime Script TM RT reagent kit (Takara, Dalian, China). The qRT-PCR was performed using SYBR® Premix Ex Taq™ II Kit (Takara). Relative expression of AC074117.1 was analyzed by 2−ΔΔCt method and normalized by GAPDH. The sequences of primers were as following: AC074117.1-forward 5ʹ-TAGGGTACACAAGGTGATCTGC-3ʹ, AC074117.1-reverse 5ʹ-CCCCACCTCCCCCAATACTA-3ʹ. GAPDH-forward 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ, GAPDH-reverse 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ.
Chromatin Immunoprecipitation (ChIP)
ChIP kit (Sigma-Aldrich, Missouri, USA) was used for ChIP assay. A549 and H1299 cells were cross-linked with 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. Cells were lysed with lysis buffer, and then sonicated for 30 min to generate DNA fragments with size of 100–1000 bp. Then, cells were immunoprecipitated with anti-H3K27ac (1:200, #4353, Cell Signaling, MA, USA) or anti-IgG (1:200, #3900, Cell Signaling). The purified DNA fragments were quantified by qRT-PCR. The primers of SEs for AC074117.1 were as follows: E1, forward: 5ʹ-CAGCATCCCAAGGCAGAAGA-3ʹ, reverse: 5ʹ-GTGTACCCAACAGCTCCGAA-3ʹ. E2, forward: 5ʹ-CAAAAACCAGTCAGGCGTGG-3ʹ, reverse: 5ʹ- TCCTCCCTCTCCCTTCTTCG-3ʹ.
Cell Proliferation Analysis
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was performed for cell proliferation analysis using cell growth determination kit (MTT based) (Sigma-Aldrich). A549 and H1299 cells with or without AC074117.1 silencing were treated with MTT (10 μL) for 5 h. Then, the cells were incubated with DMSO (150 μL) for 10 min. Absorbance at 450 nm was detected by applying a microplate reader (Beckman Coulter, CA, USA).
Luciferase Activity Assay
The candidate targets binding sites of AC074117.1, FKBP3 or E2F2 with hsa-let-7c-5p were selected using starBase v2.0. pGL3-basic vector was purchased from Promega (Fitchburg, USA). Construction of recombinant vectors containing 50 bp each upstream and downstream of the wild-type (WT) or mutant (MUT) target sites of AC074117.1, FKBP3 or E2F2 were carried out by GenePharma (Shanghai, China). AC074117.1, FKBP3 and E2F2 sequences containing wild-type (WT) or mutant (MUT) hsa-let-7c-5p binding sites are shown in Supplementary Table 1. A549 and H1299 cells were seeded into 24-well plates and cultured for 24 h before transfection. The recombined vectors and hsa-let-7c-5p mimics or control mimics (NC mimics) were co-transfected into A549 and H1299 cells using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured by Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla activity.
Statistical Analysis
Pearson’s correlation analysis was applied to screen the overlapping co-expressed genes of RNAs in the prognostic ceRNA sub-network. Pearson’s correlation between the expression of genes in the prognostic ceRNA sub-network and DNA mismatch repair markers (MLH1, EPCAM, PMS2, MSH6 and MSH2) was analyzed by R. One-way analysis of variance followed by post-hoc Tukey’s tests was applied for comparison among more than two groups. P<0.05 was considered as statistically significant.
Results
Construction of DERNAs-Based ceRNA Network in Metastatic LUAD
Compared with the adjacent normal tissues, 2355 DEmRNA, 483 DElncRNA and 155 DEmiRNA were identified in tumor tissues of patients with metastatic LUAD. The top five up- and down-regulated mRNAs, lncRNA and miRNA are displayed in Figure 1A. Subsequently, all of the DEmRNA, DElncRNA and DEmiRNA were used to construct ceRNA network. As shown in Figure 1B, a ceRNA network consisted of 40 nodes (including 7 lncRNAs, 18 miRNAs and 15 mRNAs) and 51 edges (including 33 miRNA-mRNA and 18 lncRNA-miRNA pairs) was constructed.
Identification of SEs and lncRNAs Regulated by SEs in LUAD Cells
Enhancers of HSC4 and A549 cells were ranked according to enhancer score. As shown in Figure 2A, 386 and 717 SEs were screened in HSC4 and A549 cells, respectively. SE-associated genes in HSC4 (n=386) and A549 (n=717) cells were identified (Figure 2B). A total of 133 SE-associated genes were overlapped between HSC4 and A549 cells (Figure 2B). Among 133 overlapping SE-associated genes, 69.92% were protein coding genes, 27.07% were ncRNAs and 3.01% were other genes (Figure 2C).
Next, H3K27ac signal at seven lncRNAs locus in the ceRNA network was analyzed based on GSE143653 dataset. As shown in Figure 3A–G, only AC074117.1 was an SE-associated lncRNA, while no SE was found around the other six lncRNAs locus on the genome. Furthermore, we verified the SEs of AC074117.1 in A549 and H1299 cells by ChIP-qPCR (Figure 3H–I).
Identification of Prognostic Biomarkers in ceRNA Network
Survival analysis of 40 genes in the ceRNA network was performed by Kaplan–Meier curves. Seven mRNAs showed significant influence on prognosis, among which high expression of GPI, PLK4, LMNB2, FKBP3, E2F2 and E2F8 was associated with an unfavorable prognosis (Figure 4A–F), while high expression of IFT57 was associated with a favorable prognosis (Figure 4G). One lncRNA (AC074117.1) and one miRNA (hsa-let-7c-5p) were identified as independent predictors for LUAD (Figure 4H–I).
 |
Figure 4 Kaplan–Meier curves of genes with significant influence on prognosis in ceRNA network. A total of seven mRNAs (A–G), one lncRNA (H) and one miRNA (I) were identified as prognostic factors. |
Subsequently, we analyzed the targeting relationships of the above nine RNAs (including seven mRNAs, one lncRNA and one miRNAs), and plotted the prognostic ceRNA sub-network. FKBP3, E2F2, AC074117.1 and hsa-let-7c-5p were enriched in a prognostic ceRNA sub-network regulated by SEs (Figure 5A). Therefore, we focused on the above four RNAs in the further study.
Risk scores were calculated based on the 4 RNAs. Patients were stratified into high- (n=236) and low-risk (n=236) groups (Figure 5B). The survival time of patients was shown in Figure 5C. The expression level of hsa-let-7c-5p in high-risk group was lower than that in low-risk group, while FKBP3, E2F2 and AC074117.1 were highly expressed in high-risk group (Figure 5D). The median survival time of high-risk group was significantly shorter than low-risk group (Figure 5E). Time-dependent ROC curves showed good prognostic accuracy (Figure 5F).
Co-Expression and Functional Analysis of the Prognostic ceRNA Sub-Network Regulated by SEs
Co-expression patterns showed that there were 1099, 2400, 1704 and 392 mRNAs co-expressed with FKBP3, E2F2, AC074117.1 and hsa-let-7c-5p, respectively (Figure 6A). A total of 33 overlapping mRNAs co-expressed with FKBP3, E2F2, AC074117.1 and hsa-let-7C-5p (Figure 6A). GO analysis indicated that the functions of 33 overlapping genes were mainly related to cell division (Figure 6B). The significant enriched KEGG pathways of the 33 genes are displayed in Figure 6C, including Fanconi anemia pathway, homologous recombination and cell cycle.
PPI Analysis of the Overlapping Co-Expressed Genes
PPI network was constructed based on the 33 overlapping genes. Among them, 29 genes were enriched in the PPI network through 159 edge connection, indicating that these overlapping genes were functionally closely related (Figure 7).
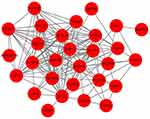 |
Figure 7 PPI network of the overlapping co-expressed genes. |
Correlation Analysis of Genes in the SE-Regulated Prognostic ceRNA Sub-Network
To further explore the relationship between the ceRNA sub-network and cell division, we analyzed the correlation between the expression of genes in the ceRNA sub-network and DNA mismatch repair markers. As shown in Figure 8, the expression levels of FKBP3, E2F2 and AC074117.1 were positively correlated with MLH1, EPCAM, PMS2, MSH6 and MSH2. Hsa-let-7c-5p was negatively correlated with EPCAM, PMS2, MSH6 and MSH2, but positively correlated with MLH1 (Figure 8).
 |
Figure 8 Pearson correlation analysis of genes expression in the SE-regulated prognostic ceRNA sub-network and DNA mismatch repair markers. |
The Effect of AC074117.1 on Cell Proliferation in LUAD Cells
Subsequently, AC074117.1 expression levels in a normal human bronchial epithelial cell line (BEAS-2B) and six LUAD cell lines (H460, HCC827, A549, H1299, PC9 and Calu3) were measured. AC074117.1 was strongly expressed in LUAD cells, especially in A549 and H1299 cells (Figure 9A). AC074117.1 was downregulated in A549 and H1299 cells transfected with sh-AC074117.1#1 or sh-AC074117.1#2 (Figure 9B). AC074117.1 silencing obviously inhibited cell proliferation in A549 and H1299 cells (Figure 9C–D).
Confirmation of the Direct Targeting Relationship of Genes in “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2”
Relative luciferase activity was notably decreased in A549 and H1299 cells co-transfected with AC074117.1-WT vector and hsa-let-7c-5p mimics, while the decrease did not occur in cells co-transfected with AC074117.1-MUT vector and hsa-let-7c-5p mimics (Figure 10A). Additionally, luciferase activity was significantly declined with FKBP3-WT vector and hsa-let-7c-5p mimics co-transfection, or E2F2-WT vector and hsa-let-7c-5p mimics co-transfection in A549 and H1299 cells (Figure 10B and C). Hence, hsa-let-7c-5p directly sponged by AC074117.1, and directly bound to FKBP3 and E2F2.
Discussion
The mortality in patients with metastatic LUAD is extremely high.1 Targeted inhibition of tumor metastasis plays important roles in improving the prognosis of advanced patients. Studies have shown that DElncRNAs are closely related to the occurrence and development of LUAD, indicating that these DElncRNAs may serve as effective biomarkers for early diagnosis and prognosis monitoring of LUAD.10,18,19 LncRNAs bind to miRNAs to regulate miRNA-mediated target genes silencing.10,11 CeRNA mechanism is one of the important mechanisms by which lncRNAs regulate proliferation, invasion, metastasis and epithelial-mesenchymal transition of LUAD cells.19–21 In this study, a ceRNA regulatory network for metastatic LUAD was initially constructed based on TCGA database.
In addition, we analyzed the SEs of LUAD cells and screened 133 SE-associated genes. SEs promote tumor progression by abnormally activating oncogenes.22 Genes regulated by SEs may be protein-coding genes or non-coding genes.17,23 Among 133 SE-associated genes in this study, 69.92% were protein-coding genes and 27.07% were non-coding RNAs. LncRNAs sponge miRNAs, thus advancing the transcription of miRNA-target genes. Therefore, we analyzed the SEs of seven lncRNAs locus in the ceRNA network, and found that only AC074117.1 was regulated by SEs. Furthermore, ChIP-qPCR was used to verify the SEs at AC074117.1 locus. These results suggested that AC074117.1 may play a key regulatory role in the entire ceRNA network.
To further reveal the biological functions of the ceRNA network constructed in this study, seven mRNAs (GPI, PLK4, LMNB2, FKBP3, E2F2, E2F8 and IFT57), one lncRNA (AC074117.1) and one miRNA (hsa-let-7c-5p) related to the overall survival of patients with metastatic LUAD were filtrated out. GPI has been confirmed to be related to high metastatic potential of glioblastoma.24 PLK4 is mainly involved in the regulation of centrosome replication.25 Overexpression of PLK4 induces tumor cell centrosome enlargement, chromosomal instability and enhances tumor invasiveness.25–27 The mammalian transcription factor E2F family is located downstream of the cell cycle signal cascade, and affects the cell cycle by regulating genes associated with mammalian cell cycle progression.28–31 E2F2 and E2F8 are oncogenes for various cancer, such as lung cancer, osteosarcoma and glioma.28–31 LMNB2, FKBP3 and IFT57 have also been confirmed to regulate cancer progression.32–34 It has been reported that hsa-let-7c-5p acts as a tumor suppressor by inhibiting proliferation and promoting apoptosis in breast cancer cells.35 At present, the role of lncRNA AC074117.1 in LUAD has not been reported. The biological functions and molecular mechanisms of AC074117.1 in LUAD need further study. Our results found that “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2” sub-network conformed to ceRNA regulatory mechanism and regulated by SEs. This SE-regulated prognostic ceRNA sub-network can be used as a potential prognostic signature for metastatic LUAD. These results further illustrated that AC074117.1 had a key regulatory role in the entire ceRNA network.
In order to further reveal the role of “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2” sub-network in metastatic LUAD, we analyzed the genes co-expressed with the prognostic ceRNA sub-network in metastatic LUAD, and performed functional cluster analysis. The overlapping co-expressed genes of AC074117.1, hsa-let-7c-5p, FKBP3 and E2F2 were mainly enriched in Fanconi anemia pathway, homologous recombination and cell cycle. Fanconi anemia pathway is essential for repairing DNA cross-linking damage and maintaining genome stability, and is therefore closely related to cancer occurrence and development.36 In addition, Fanconi anemia pathway is related to DNA homologous recombination during the division phase of cell cycle and cell cycle checkpoints.37 The reduction of DNA repair ability caused by Fanconi anemia pathway dysfunction leads to increase in the probability of tumors, and increases tumor chemosensitivity that affects DNA cross-linking.38,39
To verify the relationship between the “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2” sub-network and DNA damage, we analyzed the correlation between the sub-network and DNA mismatch repair markers. Our results found that “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2” sub-network was significantly correlated with DNA mismatch repair. Therefore, we speculated that “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2” sub-network promotes LUAD metastasis by regulating the repair of DNA damage. To our best knowledge, the function and regulation of AC074117.1 in LUAD have not been reported. In this study, we verified the expression level of AC074117.1 in LUAD cells was higher than that in the normal human bronchial epithelial cell, and discovered that AC074117.1 silencing inhibited LUAD cell proliferation. Furthermore, we confirmed the direct binding relationships in “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2” sub-network. However, this study has some limitations. The functions of other genes in the network except for AC074117.1 were not explored in this study, which will be further explored in our following studies.
In conclusion, by constructing ceRNA network in metastatic LUAD, we screened out seven mRNA, one lncRNA and one miRNA as novel candidate prognostic factors for metastatic LUAD. Among them, “AC074117.1-hsa-let-7c-5p-FKBP3/E2F2” is a key ceRNA sub-network regulated by SEs. These results provide potential therapeutic targets and prognostic-related molecular markers for further study of metastatic LUAD progression.
Data Sharing Statement
The data in this study can be obtained under reasonable conditions by contacting Dr. Weidong Zhang (email: [email protected]).
Funding
This study was supported by the Tianjin First Central Hospital Science and Technology Fund Project (2019CL02).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zhang H, Guo L, Chen J. Rationale for lung adenocarcinoma prevention and drug development based on molecular biology during carcinogenesis. Onco Targets Ther. 2020;13:3085–3091. doi:10.2147/OTT.S248436
2. Sacher AG, Dahlberg SE, Heng J, Mach S, Janne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol. 2016;2(3):313–320. doi:10.1001/jamaoncol.2015.4482
3. Huang J, Pan B, Xia G, Zhu J, Li C, Feng J. LncRNA SNHG15 regulates EGFR-TKI acquired resistance in lung adenocarcinoma through sponging miR-451 to upregulate MDR-1. Cell Death Dis. 2020;11(7):525. doi:10.1038/s41419-020-2683-x
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
5. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
6. Sedano MJ, Harrison AL, Zilaie M, et al. Emerging roles of estrogen-regulated enhancer and long non-coding RNAs. Int J Mol Sci. 2020;21(10):3711. doi:10.3390/ijms21103711
7. Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217. doi:10.3727/096504016X14549667334007
8. Manning AK, Goustin AS, Kleinbrink EL, et al. A long non-coding RNA, LOC157273, is an effector transcript at the chromosome 8p23.1-PPP1R3B metabolic traits and type 2 diabetes risk locus. Front Genet. 2020;11:615. doi:10.3389/fgene.2020.00615
9. Collins L, Binder P, Chen H, Wang X. Regulation of long non-coding RNAs and microRNAs in heart disease: insight into mechanisms and therapeutic approaches. Front Physiol. 2020;11:798. doi:10.3389/fphys.2020.00798
10. Zhao X, Li X, Zhou L, et al. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci. 2018;109(10):3068–3079. doi:10.1111/cas.13764
11. Chen R, Xia W, Wang S, et al. Long noncoding RNA SBF2-AS1 is critical for tumorigenesis of early-stage lung adenocarcinoma. Mol Ther Nucleic Acids. 2019;16:543–553. doi:10.1016/j.omtn.2019.04.004
12. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
13. Liu X, Huang S, Guan Y, Zhang Q. Long noncoding RNA OSER1AS1 promotes the malignant properties of nonsmall cell lung cancer by sponging microRNA4333p and thereby increasing Smad2 expression. Oncol Rep. 2020;44(2):599–610. doi:10.3892/or.2020.7645
14. Liu C, Li X, Hao Y, et al. STAT1-induced upregulation of lncRNA KTN1-AS1 predicts poor prognosis and facilitates non-small cell lung cancer progression via miR-23b/DEPDC1 axis. Aging. 2020;12(9):8680–8701. doi:10.18632/aging.103191
15. Fang C, Wang L, Gong C, Wu W, Yao C, Zhu S. Long non-coding RNAs: how to regulate the metastasis of non-small-cell lung cancer. J Cell Mol Med. 2020;24(6):3282–3291. doi:10.1111/jcmm.15054
16. Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi:10.1016/j.cell.2013.03.035
17. Li GH, Qu Q, Qi TT, et al. Super-enhancers: a new frontier for epigenetic modifiers in cancer chemoresistance. J Exp Clin Cancer Res. 2021;40(1):174. doi:10.1186/s13046-021-01974-y
18. Yang D, He Y, Wu B, et al. Predictions of the dysregulated competing endogenous RNA signature involved in the progression of human lung adenocarcinoma. Cancer Biomark. 2020;29(3):399–416. doi:10.3233/CBM-200133
19. Zhang X, Du L, Han J, et al. Novel long non-coding RNA LINC02323 promotes epithelial-mesenchymal transition and metastasis via sponging miR-1343-3p in lung adenocarcinoma. Thorac Cancer. 2020;11(9):2506–2516
20. Zhao M, Xin XF, Zhang JY, Dai W, Lv TF, Song Y. LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. Cancer Med. 2020;9(3):1196–1208. doi:10.1002/cam4.2776
21. Jia Y, Duan Y, Liu T, et al. LncRNA TTN-AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging miR-142-5p to regulate CDK5. Cell Death Dis. 2019;10(8):573. doi:10.1038/s41419-019-1811-y
22. Thandapani P. Super-enhancers in cancer. Pharmacol Ther. 2019;199:129–138. doi:10.1016/j.pharmthera.2019.02.014
23. Sengupta S, George RE. Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer. 2017;3(4):269–281. doi:10.1016/j.trecan.2017.03.006
24. Kathagen-Buhmann A, Maire CL, Weller J, et al. The secreted glycolytic enzyme GPI/AMF stimulates glioblastoma cell migration and invasion in an autocrine fashion but can have anti-proliferative effects. Neuro Oncol. 2018;20(12):1594–1605. doi:10.1093/neuonc/noy117
25. Maniswami RR, Prashanth S, Karanth AV, et al. PLK4: a link between centriole biogenesis and cancer. Expert Opin Ther Targets. 2018;22(1):59–73. doi:10.1080/14728222.2018.1410140
26. Zhu Y, Liu Z, Qu Y, et al. YLZ-F5, a novel polo-like kinase 4 inhibitor, inhibits human ovarian cancer cell growth by inducing apoptosis and mitotic defects. Cancer Chemother Pharmacol. 2020;86(1):33–43. doi:10.1007/s00280-020-04098-w
27. Shinmura K, Kurabe N, Goto M, et al. PLK4 overexpression and its effect on centrosome regulation and chromosome stability in human gastric cancer. Mol Biol Rep. 2014;41(10):6635–6644. doi:10.1007/s11033-014-3546-2
28. Yu H, Zhang D, Li Z, Wang M. E2F transcription factor 8 promotes proliferation and radioresistance in glioblastoma. Pathol Res Pract. 2020;216(8):153030. doi:10.1016/j.prp.2020.153030
29. Yan Z, Zhang W, Xiong Y, Wang Y, Li Z. Long noncoding RNA FLVCR1-AS1 aggravates biological behaviors of glioma cells via targeting miR-4731-5p/E2F2 axis. Biochem Biophys Res Commun. 2020;521(3):716–720. doi:10.1016/j.bbrc.2019.10.106
30. Iwasaki T, Tanaka K, Kawano M, Itonaga I, Tsumura H. Tumor-suppressive microRNA-let-7a inhibits cell proliferation via targeting of E2F2 in osteosarcoma cells. Int J Oncol. 2015;46(4):1543–1550. doi:10.3892/ijo.2015.2867
31. Dou L, Han K, Xiao M, Lv F. miR-223-5p suppresses tumor growth and metastasis in non-small cell lung cancer by targeting E2F8. Oncol Res. 2019;27(2):261–268. doi:10.3727/096504018X15219188894056
32. Misono S, Seki N, Mizuno K, et al. Dual strands of the miR-145 duplex (miR-145-5p and miR-145-3p) regulate oncogenes in lung adenocarcinoma pathogenesis. J Hum Genet. 2018;63(10):1015–1028. doi:10.1038/s10038-018-0497-9
33. Tong J, Shen Y, Chen X, et al. FKBP3 mediates oxaliplatin resistance in colorectal cancer cells by regulating HDAC2 expression. Oncol Rep. 2019. doi:10.3892/or.2019.7259
34. Gdynia G, Lehmann-Koch J, Sieber S, et al. BLOC1S2 interacts with the HIPPI protein and sensitizes NCH89 glioblastoma cells to apoptosis. Apoptosis. 2008;13(3):437–447. doi:10.1007/s10495-007-0176-3
35. Fu X, Mao X, Wang Y, Ding X, Li Y. Let-7c-5p inhibits cell proliferation and induces cell apoptosis by targeting ERCC6 in breast cancer. Oncol Rep. 2017;38(3):1851–1856. doi:10.3892/or.2017.5839
36. Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17(6):337–349. doi:10.1038/nrm.2016.48
37. Kolinjivadi AM, Crismani W, Ngeow J. Emerging functions of Fanconi anemia genes in replication fork protection pathways. Hum Mol Genet. 2020;29(R2):R158–R164. doi:10.1093/hmg/ddaa087
38. Roh YG, Mun JY, Kim SK, et al. Fanconi anemia pathway activation by FOXM1 is critical to bladder cancer recurrence and anticancer drug resistance. Cancers. 2020;12(6):1417. doi:10.3390/cancers12061417
39. Fang CB, Wu HT, Zhang ML, Liu J, Zhang GJ. Fanconi anemia pathway: mechanisms of breast cancer Predisposition development and potential therapeutic targets. Front Cell Dev Biol. 2020;8:160. doi:10.3389/fcell.2020.00160
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.