Back to Journals » Journal of Inflammation Research » Volume 16
Identification of Key Immune-Related Genes in the Treatment of Heart Failure After Myocardial Infarction with Empagliflozin Based on RNA-Seq
Authors Zhang P, Wang TY, Luo ZY , Ding JC, Yang Q , Hu PF
Received 3 July 2023
Accepted for publication 10 October 2023
Published 18 October 2023 Volume 2023:16 Pages 4679—4696
DOI https://doi.org/10.2147/JIR.S428747
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Pei Zhang,1,* Tian-Yu Wang,2,* Zi-Yue Luo,2 Jun-Can Ding,2 Qiang Yang,2 Peng-Fei Hu3
1Department of Cardiology, Sir Run Run Shaw Hospital, College of Medicine Zhejiang University, Hangzhou, Zhejiang Province, 310018, People’s Republic of China; 2Second Clinical Medical College, Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310053, People’s Republic of China; 3Department of Cardiology, the Second Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310005, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Peng-Fei Hu, Department of Cardiology, The Second Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310005, People’s Republic of China, Tel +86 15267037741, Email [email protected]
Purpose: Heart failure is a serious complication after acute myocardial infarction (AMI). It is crucial to investigate the mechanism of action of empagliflozin in the treatment of heart failure.
Methods: A total of 20 wild type (WT) male C57BL6/J mice were used to establish a model of heart failure after myocardial infarction and randomly divided into 2 groups: treatment group and control group. The treatment group was treated with empagliflozin, and the control group was treated with placebo. After 8 weeks of treatment, mouse heart tissues were collected for next generation sequencing. Bioinformatics methods were used to screen the key genes. Finally, the correlation between clinical data and gene expression was analyzed. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to verify the expression of key genes.
Results: A mouse model of heart failure was successfully constructed. By DEG analysis, a total of 740 DEGs in the treatment group vs the control group were obtained. Dendritic cells, granulocytes, follicular B, plasma cell, cDC1, cDC2, pDC and neutrophils were 8 different immune cells identified by immunoinfiltration analysis. Through WGCNA, the turquoise module with the highest correlation with the above differential immune cells was selected. One hundred and forty-two immune-related DEGs were obtained by taking intersection of the DEGs and the genes of the turquoise module. Col17a1 and Gria4 were finally screened out as key immune-related genes via PPI analysis and machine learning. Col17a1 was significantly up-regulated, while Gria4 was significantly down-regulated in the treatment group. At the same time, the expression level of Col17a1 was significantly correlated with left ventricular ejection fraction (LVEF), left ventricular fraction shortening (LVFS) and left ventricular internal dimension systole (LVIDs).
Conclusion: Col17a1 and Gria4 are key immune-related genes of empagliflozin in the treatment of heart failure after myocardial infarction. This study provides a scientific basis for elucidating the mechanism of action of empagliflozin in treating heart failure after myocardial infarction.
Keywords: heart failure after myocardial infarction, empagliflozin, immunology, RNA-seq
Introduction
Heart failure is caused by blood stasis in the venous system and insufficiency of blood perfusion in the arterial system due to the dysfunction of systolic or diastolic function of the heart, and it is the terminal stage for almost all kinds of heart diseases.1 Acute myocardial infarction (AMI) is myocardial necrosis caused by acute and persistent ischemia and hypoxia in coronary arteries, and heart failure is one of the most common and harmful complications after AMI.2,3 Recent epidemiological reports show that about 20–30% of patients are diagnosed with heart failure 1 year after AMI, and complicated heart failure increases the overall risk of death by 3 times and cardiovascular mortality by 4 times.4 Therefore, heart failure after myocardial infarction is seriously endangering human health and life safety.
Since AMI is one of the most important etiologies of heart failure, an increasing number of studies have focused on exploring the mechanism of heart failure after myocardial infarction. Immuno-inflammatory response plays an important role in the occurrence and development of heart failure after myocardial infarction.5 For example, cardiomyocyte necrosis has been reported to be one of the factors causing heart failure after myocardial infarction, and the inflammatory responses caused by the death of cardiomyocytes can further aggravate the progression of heart failure after myocardial infarction.6,7 Studies have also confirmed that many immune cells, including neutrophils and lymphocytes, can affect inflammation and ventricular remodeling after AMI, and ultimately lead to heart failure.8,9 In addition, a retrospective cohort study showed that a high level of systemic immunoinflammatory index often represents a worse prognosis for patients with heart failure.10 Undeniably, immunity greatly affects the progression of heart failure after myocardial infarction.
Empagliflozin is a sodium-dependent glucose transporters-2 (SGLT-2) inhibitor originally developed for the treatment of type 2 diabetes and has been approved in the European Union and the United States for the treatment of chronic heart failure.11 A large number of clinical studies have shown that empagliflozin can significantly reduce the combined risk of cardiovascular death in heart failure patients with reduced or preserved ejection fraction,12,13 and reduce obesity-related chronic inflammation and insulin resistance by reducing the accumulation of M1-polarized macrophages.14 In addition, empagliflozin significantly improves the liver injury associated with non-alcoholic fatty liver disease by inhibiting the inflammatory response mediated by the IL-17/IL-23 axis.15 In terms of cardiovascular protection, studies have found that empagliflozin can reduce inflammation in patients with heart failure with preserved ejection fraction (HFpEF), thereby improving their cardiomyocyte function.16 However, the potential mechanism of empagliflozin in the treatment of heart failure after myocardial infarction remains to be further elucidated.
With the rapid development of second-generation sequencing technology and bioinformatics, a growing number of studies have begun to explore gene targets related to drug therapy from the gene level, so as to continuously improve the relevant molecular mechanisms of related drugs in disease treatment.17 For example, transcriptome sequencing has shown that empagliflozin can target activation of the AKT/eNOS/NO pathway, thereby contributing to the inhibition of endothelial cell apoptosis, maintenance of capillary formation, and improvement of systolic dysfunction during left ventricular pressure overload.18 In this study, a mouse model of myocardial infarction was constructed by ligation of the left anterior descending branch of the experimental mice, and echocardiography was carried out 3 days after myocardial infarction to verify the model establishment of heart failure after myocardial infarction.19 The mice were randomly divided into 2 groups: the treatment group and the control group, and treated with empagliflozin or placebo, respectively, for 8 weeks, after which their myocardial tissues were collected for RNA-sequencing (RNA-seq). Then, bioinformatic analysis was performed on the sequencing data to identify key immune-related genes in mice with heart failure after myocardial infarction treated with empagliflozin.
Methods
Breeding of Laboratory Animals
Twenty healthy male C57BL6/J mice aged 6–8 weeks were raised in Zhejiang Center of Laboratory Animals (ZJCLA). All mice were adaptively fed for 1 week with (45 ± 5) % humidity, (22 ± 2)°C temperature, 12h/12h cycle of light and darkness, free access to drinking water, and ordinary feed. This animal research protocol was approved by Institutional Animal Care and Use Committee, ZJCLA (Approval No. ZJCLA-IACUC-20120005).
Establishment of Animal Models
After 1 week of adaptive feeding, all the mice were subjected to ligation of the left anterior descending coronary artery for model establishment of heart failure after myocardial infarction. The mice were anesthetized by intraperitoneal injection of 0.5% pentobarbital sodium (0.08mL/10g), and their hearts were exposed by thoracotomy at the third and fourth intercostal ribs of the left chest. The left anterior descending coronary artery was ligated with 7–0 operating line 1mm below the left atrial appendage. After part of the myocardium turned white, the rib, muscle and epidermis of the mice were sutured layer by layer with the 5–0 operating line. Post-operative care was given to the mice until their recovery. The survival, food intake, mental state, and activity of mice were observed 3 days after myocardial infarction. If the mice developed symptoms of emaciation, depilation, mental fatigue, and decreased activity, and the left ventricular ejection fraction (LVEF) ≤60% as determined by echocardiography, the successful mouse model establishment of heart failure after myocardial infarction was indicated.20 The 20 mice were randomly divided into two groups: the treatment group and the control group. Mice in the treatment group were treated with empagliflozin at a dose of 10mg/kg/d and those in the control group was given placebo at the same dosage. After 8 weeks of treatment, echocardiography was used to detect the cardiac functions. All the mice were euthanized, and RNA was extracted from their myocardial tissues.
RNA-Seq
The Agilent 2100 bioanalyzer was used to accurately detect the integrity and total amount of RNA in the mouse myocardial tissues. The starting RNA for library construction was total RNA, and mRNA with polyA tails was enriched by Oligo(dT) magnetic beads. The resulting mRNA was then randomly interrupted with divalent cations in Fragmentation Buffer. The fragmented mRNA was used as a template and random oligonucleotides were used as primers to synthesise the first strand of cDNA in the M-MuLV reverse transcriptase system, followed by degradation of the RNA strand with RNaseH and synthesis of the second strand of cDNA with dNTPs in the DNA polymerase I system. The purified double-stranded cDNAs were end-repaired, A-tailed and sequenced. The cDNAs of 370–420 bp were screened for PCR amplification using AMPure XP beads, and the PCR products were purified again using AMPure XP beads to obtain the final library. After library construction, the library was initially quantified using a Qubit 2.0 Fluorometer and diluted to 1.5 ng/ul, followed by an Agilent 2100 bioanalyzer to check the insert size of the library. After the library size met the expectations, qRT-PCR was performed to accurately quantify the effective concentration of the library (effective library concentration above 2 nM) to ensure the quality of the library. Once the libraries have passed the test, the libraries were pooled according to the effective concentration and the target downstream data volume and then sequenced by Illumina NovaSeq 6000 to produce 150 bp paired end reads. Four fluorescently labelled dNTPs, DNA polymerase and splice primers were added to the sequencing flow cell for amplification.
Detection of the Cardiac Function of Mice by Echocardiography
The left ventricular internal dimension diastole (LVIDd), left ventricular internal dimension systole (LVIDs), LVEF, left ventricular fractional shortening (LVFS) of mice in each group were measured using the small animal ultrasound imaging system systole. The cardiac ultrasound data of mice were recorded before surgery, 3 days and 8 weeks after surgery, respectively.
RNA-Seq and Data Processing
The sequencing data were collected from the myocardial tissues of the 20 mice, including 10 control samples and 10 empagliflozin-treated samples. Illumina NovaSeq 6000 was used for double-ended PE150 sequencing, and fastp (version 0.23.2) was used for data quality control and filtering. Then, hisat2 (version 2.2.1) was used for sequence alignment with mouse GRCm39 as the reference genome, and samtools (version 1.15.1) was used to convert the sam format file into bam format file. Afterwards, featureCounts (version 2.0.1) was used to recount the count gene expression profiles of all the samples from the annotated gtf file of the mouse GRCm39. The amount of sequencing data is shown in Table S1.
Differentially Expressed Genes (DEGs) Analysis and Gene Set Enrichment Analysis (GSEA)
The edgeR of R package (version 3.36) was used to screen DEGs in the treatment group vs the control group according to |logFC|>1 and P value < 0.05. Volcano mapping was subsequently performed on the DEGs, and DEGs thermal mapping was analyzed using the pheatmap package (version 1.0.12). GSEA software (version 4.3.2) was used for GSEA, and significantly enriched pathways were obtained in the treatment group.
Immunoinfiltration Analysis
Immunoinfiltration analysis of the expression data was performed using the ImmuCellAI-mouse database (http://bioinfo.life.hust.edu.cn/ImmuCellAI-mouse/#! /). The immunoinfiltration results were visualized using the R package, and the corresponding infiltration histogram was plotted. In addition, heat maps of intercellular correlations were plotted using the corrplot package (version 0.92). Intergroup expression differences were analyzed according to the Wilcoxon test, and violin plots of component differences were plotted using the vioplot package (version 0.3.7).
Weighted Gene Co-Expression Network Analysis (WGCNA)
WGCNA was performed using R’s WGCNA package (version 1.71) on the standardized presentation data. Firstly, the samples were clustered, and heat maps of the sample clustering and clinical characteristics were drawn. Independence and mean connectivity analyses were then performed to obtain power values. The weighted co-expression network was constructed according to the power values. The correlation analysis of the sample characteristics and modules was carried out, and a correlation heat map was drawn.
Key Gene Screening and Functional Enrichment Analysis
Genes in the key modules of WGCNA were interested with DEGs to obtain key genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the clusterProfiler package of R (version 4.2.2).
Protein–Protein Interaction (PPI) Analysis
STRING (version 11.5, https://cn.string-db.org/) was used for PPI analysis of the key genes. The PPI analysis results were imported into Cytoscape (version 3.9.1) and analyzed using cytohubba plug-in. Degree, DMNC, EPC, MCC and MNC were algorithms, respectively, used to get the top 50 genes, and the results obtained were plotted on an upset graph.
Machine Learning Analysis
The genes obtained after PPI analysis were analyzed by machine learning. The glmnet package (version 4.1–4) was used for lasso analysis, the randomForest package (version 4.7–1.1) was used for random forest analysis, and the e1071 package (version 1.7–11) and the caret package (version 6.0–92) were used for SVM-RFE analysis. The results via the above three machine learning methods were interested, and interested genes were obtained.
Receiver Operating Characteristic (ROC) Curve Analysis and Correlation Analysis
Boxmaps of the key gene expression were drawn, and ROC analysis was performed using the pROC package (v 1.18). The R software was used to analyze the clinical data. According to the treatment group vs the control group, boxplot analysis and graph drawing were performed using the ggpubr package (v 0.4) and the ggplot2 package (v 3.3.6) by t-test. The ggExtra package (v 0.10) and the stats package (v 4.2.1) were used for correlation analysis and mapping of the clinical data, immune cells and gene expression by spearman. A lollipop chart was drawn based on the results of correlation analysis.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNA was collected from the heart tissues and converted into cDNA by reverse transcription using the Evo M-MLV RT Premix for qPCR kit. The relative expression levels of Col17a1 and Gria4 were detected using the HieffqPCR SYBRGreen Master Mix (No Rox) kit. The primer sequences are provided in Table 1, and specific experimental procedures can be found in the kit manual.
 |
Table 1 Primer Sequence Information in the Experiment |
Results
Establishment of a Mouse Model of Heart Failure After Myocardial Infarction
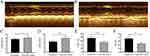
The LVIDd, LVIDs, LVEF and LVFS of the 20 healthy male C57BL6/J mice were detected by echocardiography before surgery (Figure 1A) and 3 days after the successful modeling of myocardial infarction by ligation of the left anterior descending coronary artery (Figure 1B). The results showed that the cardiac ultrasound parameters were better before surgery than 3 days after surgery, and the LVEF was lower than 60% at 3 days after surgery, suggesting that a mouse model of heart failure after myocardial infarction was successfully established (Figure 1C–F).
Screening and Enrichment Analysis of DEGs
DEGs in the treatment group vs the control group were screened according to a threshold of |logFC|>1 & P value < 0.05 and plotted into the volcano map (Figure 2A) and heat map (Figure 2B). Differential analysis showed that a total of 740 DEGs were screened out between groups, with 425 up-regulated and 315 down-regulated. GSEA results showed that genes were significantly enriched in pathways related to immune inflammation in the treatment group, such as the Toll pathway, AKT pathway, IL-9 signaling pathway and IL-6 signaling pathway (Figure 3A–D).
 |
Figure 2 Screening of DEGs. (A) Heatmap showing DEGs in the treatment group vs the control group. (B) Volcano map showing DEGs in the treatment group vs the control group. |
Immune Infiltration Analysis
The ImmuCellAI-mouse database was used for performing immune infiltration analysis of the expression data, and the analysis results were shown in Table S2. The R package was used to visualize the immune infiltration results, and a histogram of infiltration (Figure 4A), and a correlation heat map between immune cells were drawn, respectively (Figure 4B). According to Wilcoxon test, differential expression analysis was performed between groups, and the differences between groups were shown by a violin chart (Figure 4C). According to P value < 0.05, 8 different immune cells were obtained, including dendritic cells, granulocytes, follicular B, plasma cells, cDC1, cDC2, pDC and neutrophils.
Identification of Immune-Related Gene Modules
R’s WGCNA package was used for WGCNA based on the standardized presentation data. Firstly, the samples were clustered, and a heatmap of the sample clustering and the clinical characteristics was drawn (Figure 5A). Subsequently, the independence and average connectivity analysis were conducted, with the power value as 2 (Figure 5B), according to which the R^2 and slope were 0.98 and −4.19, respectively (Figure 5C), and a weighted co-expression network (Figure 5D) was constructed. Correlation analysis of the sample characteristics and modules was conducted, and the analysis results were shown by a heat map (Figure 6A). The turquoise module was selected as the key module according to correlation and significance (P<0.05). The correlation between the turquoise module and the immune cells was analyzed (Figure 6B), and P value < 0.001 was significantly correlated. The genes of the turquoise module were significantly correlated with dendritic cells, granulocytes, cDC1, cDC2, pDC and neutrophils.
Key Gene Analysis and Enrichment Analysis
Genes in the key modules of WGCNA were interested with DEGs, and 142 key genes were obtained (Figure 7A) and subjected to GO and KEGG enrichment analysis (Figure 7B and Table S3). These intersection genes were revealed to be significantly enriched in cartilage development involved in biological processes (BP) such as endochondral bone morphogenesis and skeletal system morphogenesis; cell components (CC) such as extracellular matrix and collagen-containing extracellular matrix; molecular functions (MF) such as extracellular matrix structural constituent and ligand-gated cation channel activity; ECM-receptor interaction, protein digestion and absorption pathways.
PPI Analysis and Machine Learning Analysis
The STRING database was used for PPI analysis of the interested key genes (Figure 8A), and Degree, DMNC, EPC, MCC and MNC were algorithms used to get the top 50 genes, respectively. The results obtained from the above five algorithms were subjected to upset analysis, and 46 genes were obtained (Figure 8B). Lasso analysis (Figure 9A and B), forest analysis (Figure 9C and D), and SVM-RFE analysis (Figure 9E) were, respectively, performed, and the machine learning analysis results obtained were interested (Figure 9F). As a result, Col17a1 and Gria4 were finally obtained and identified as key immune-related genes.
 |
Figure 8 PPI analysis and upset analysis. (A) PPI network of the 142 genes. (B) The intersected genes obtained by five algorithms shown by upset diagram. |
ROC Analysis and Correlation Analysis
The expression boxplot of the Col17a1 and Gria4 was drawn (Figure 10A and B) and ROC curve analysis (Figure 10C and D) was performed. Col17a1 was significantly highly expressed while Gria4 was significantly lowly expressed in the treatment group. The AUC area of Col17a1 and Gria4 is 0.795 and 0.860, respectively. The correlation between the two genes and the immune cells was analyzed, and a lollipop diagram was drawn (Figure 11A and B). Col17a1 was revealed to be significantly correlated with M1 macrophage, granulocytes, neutrophils, dendritic cells, B cell, memory B, cDC2, pDC, cDC1 and follicular B, while Gria4 was significantly correlated with cDC1, mast cells, neutrophils and plasma cells (Figures S1 and S2).
Analysis of the Correlation Between the Key Genes and the Clinical Data
Mice in the treatment group were administered with empagliflozin and those in the control group were given placebo. After 8 weeks of treatment, echocardiography was performed on the mice, and the echocardiographic results were compared between the treatment group and the control group. Significant differences were shown in LVIDd, LVIDs, LVEF, and LVFS between groups (Figure 12A). The correlation between the two genes (Col17a1 and Gria4) and the clinical data was analyzed, and a lollipop diagram was drawn (Figure 12B). Col17a1 was shown to be significantly correlated with LVIDs, LVEF and LVFS, but was not significantly correlated with LVIDd (Figure 12C), while no significant correlation was found between Gria4 and LVIDd, LVIDs, LVEF, and LVFS (Figure 12D).
Validation of Key Genes
To further verify the expression of key genes, we detected the expression levels of Col17a1 and Gria4 in the heart tissues of mice by qRT-PCR. The results showed that Col17a1 was significantly highly expressed in the treatment group, while Gria4 was significantly lowly expressed in the treatment group, which was consistent with the trend of the sequencing results (Figure 13A and B).
 |
Figure 13 Verification of gene expression by qRT-PCR. (A) The expression of Col17a1 by qRT-PCR. (B) The expression of Gria4 by qRT-PCR. ***P < 0.001. |
Discussion
Heart failure is the end-stage of various cardiovascular diseases. It is accompanied by changes in cardiac structure and function and characterized by high incidence, recurrence and mortality.21 AMI, as one of the most severe acute cardiovascular diseases, is easy to cause heart failure.22 Numerous studies have shown that in the pathogenesis of AMI, the damage caused by continuous myocardial ischemia and hypoxia leads to early inflammatory responses of necrotic myocardium, which further aggravates the process of ventricular remodeling and myocardial fibrosis, thus increasing the risk of heart failure.23,24 There is no doubt that heart failure after AMI is a great threat to human life and health. As a drug newly discovered for treating heart failure, empagliflozin has been proved to have good efficacy in patients with heart failure and has now been gradually widely used in clinical practice.25 However, the mechanism of its action in treating heart failure is still unclear and needs further study.
In this study, a mouse model of myocardial infarction was established by ligation of the left anterior descending coronary artery of the experimental mice, and echocardiography was carried out 3 days later in order to verify whether a model of heart failure was successfully constructed.19 Afterwards, mice in the treatment group and the control group were treated with the corresponding drugs for 8 weeks, after which their myocardial tissues were collected for RNA-seq.
A total of 740 DEGs in the treatment group and control group were screened by difference analysis. GSEA results showed that genes in the treatment group were significantly enriched in the Toll pathway, AKT pathway, IL-9 signaling pathway and IL-6 signaling pathway related to immune inflammation. Currently, a large number of studies have shown that cells in the immune system can mediate the protective and destructive effects of heart remodeling, and inflammation has become an important factor in the occurrence and development of heart failure.26 Toll-like receptors have been shown to affect the progression of heart failure after myocardial infarction by mediating the release of inflammatory cytokines.27 Calycosin has been reported to inhibit inflammation and fibrosis in rats with heart failure after myocardial infarction by activating PI3K-AKT pathway.28 In addition, both IL-9 and IL-6, as members of the interleukin family, have been proven to be major inflammatory mediators involved in heart failure.29,30 Therefore, empagliflozin may improve heart failure by mediating the above immune inflammatory signaling pathways.
Based on the above results, immunoinfiltration analysis and WGCNA were performed. Dendritic cells, granulocytes, follicular B, plasma cells, cDC1, cDC2, pDC and neutrophils were 8 different immune cells between groups shown by immunoinfiltration. cDC1, cDC2 and pDC are different subtypes of dendritic cells, and dendritic cells have also been proved to dominate various functions of the immune system.31 Besides, dendritic cells can improve the remodeling and cardiac function after myocardial infarction by regulating the polarization of regulatory T cells and macrophages,32 and advanced glycation end products (AGEs) can promote the progression of heart failure by inducing the maturation of dendritic cells.33 Neutrophils are the most abundant granulocyte type and are considered to be regulators of cardiovascular inflammation due to their important roles in cardiovascular inflammation and repair.34 Recent studies have shown that KLF2 can affect the occurrence and development of heart failure by regulating neutrophil-mediated immune thrombotic dysregulation.35 Both follicular B and plasma cells belong to the B cell family. It has been reported that B cells can affect inflammation and ventricular remodeling after AMI and eventually lead to heart failure. Therefore, B cells are gaining popularity as potential targets for the novel immunotherapy of heart failure.9,36
In order to further explore the correlation between genes and immune cells, the turquoise module significantly correlated with dendritic cells, granulocytes, cDC1, cDC2, pDC and neutrophils was selected by WGCNA in this study. A total of 142 immune-related DEGs were obtained by taking intersection of genes in the turquoise module and the DEGs. After PPI, machine learning and ROC analyses, Col17a1 and Gria4 were finally obtained as key immune-related genes. The former was significantly overexpressed in the treatment group and significantly correlated with M1 macrophage, granulocytes, neutrophils, dendritic cells, B cells, memory B, cDC2, pDC, cDC1 and follicular, while the latter was significantly underexpressed in the treatment group and significantly correlated with cDC1, mast cells, neutrophils and plasma cells. Therefore, according to the above results, it can be speculated that Col17a1 and Gria4 may ameliorate heart failure after myocardial infarction by inhibiting the release of pro-inflammatory cytokines by immune cells such as M1 macrophages, neutrophils, dendritic cells, and B cells. Col17a1 is type ⅩⅤII collagen, which can mediate the adhesion between keratinocytes and basement membrane and has been confirmed to be abnormally expressed in multiple cancers.37,38 Studies have shown that Col17a1 has a certain correlation with immune inflammation. For example, Col17a1 regulates the expression level of the pro-inflammatory factor IL-8 in epidermal keratinocytes,39 and can also inhibit the proliferation of breast cancer cells by inhibiting the activity of the AKT/mTOR signaling pathway.40 Although type ⅩⅤII collagen is poorly studied in cardiovascular diseases, many studies emphasize the potential application of type I and type III collagen as biomarkers of myocardial fibrosis and prognostic markers of heart failure.41 Gria4 encodes a member of the L-glutamate-gated ion channel family, and high levels of plasma glutamate have been shown to be associated with an increased risk of heart failure.42 In addition, mangiferin inhibits cardiac fibrosis by redistributing glutamic acid through activation of Nrf2.43 Although the mechanism of Col17a1 and Gria4 in heart failure has not been thoroughly elaborated, it can be speculated based on the above research results that empagliflozin may regulate the immune inflammatory responses by targeting Col17a1 and Gria4 so as to play certain roles in the treatment and improvement of heart failure after myocardial infarction.
In this study, the results of echocardiography were compared between groups, and the LVIDd, LVIDs, LVEF, and LVFS were significantly better in the treatment group than in the control group, confirming the efficacy of empagliflozin in the treatment of heart failure after myocardial infarction. This study further analyzed the correlation between the key genes and the clinical data, and Col17a1 was revealed to be significantly correlated with LVIDs, LVEF and LVFS, but not with the level of LVIDd, while Gria4 was not significantly correlated with LVIDd, LVIDs, LVEF and LVFS. However, it should be noted that there are limitations of this study. In the first place, the levels of other indicators except LVIDd, LVIDs, LVEF and LVFS between groups were not detected. Therefore, the clinical significance of Col17a1 and Gria4 remains to be further verified by subsequent studies. In addition, the sample size of this study is small, and there are a limited number of studies on the mouse model of heart failure after myocardial infarction, so a larger cohort of clinical trials is still needed.
Conclusion
Col17a1 and Gria4 may be the key immune-related genes of empagliflozin in the treatment of heart failure after myocardial infarction. Empagliflozin may regulate immune inflammatory responses, so as to effectively treat and improve heart failure after myocardial infarction. This study provides a reliable theoretical basis for using empagliflozin to treat heart failure induced by myocardial infarction, and also reveals the related mechanism of action during the process of treatment.
Ethics Approval and Informed Consent
The use of animal experiments was approved by the Institutional Animal Care and Use Committee. (Ethical number: ZJCLA-IACUC-20120005) All animal use, care, and operative procedures complied with the Guide for the Care and Use of Laboratory Animals by the National Institution of Health.
Funding
This study was supported by the Zhejiang Medicine and Health Science and Technology Plan Project (project number: 2020KY205) and the Zhejiang TCM Science and Technology Planning Project (project number: 2022ZA082).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Roger VL. Epidemiology of heart failure: a contemporary perspective. Circ Res. 2021;128(10):1421–1434. doi:10.1161/CIRCRESAHA.121.318172
2. Peet C, Ivetic A, Bromage DI, et al. Cardiac monocytes and macrophages after myocardial infarction. Cardiovasc Res. 2020;116(6):1101–1112. doi:10.1093/cvr/cvz336
3. Wu X, Reboll MR, Korf-Klingebiel M, et al. Angiogenesis after acute myocardial infarction. Cardiovasc Res. 2021;117(5):1257–1273. doi:10.1093/cvr/cvaa287
4. Jenča D, Melenovský V, Stehlik J, et al. Heart failure after myocardial infarction: incidence and predictors. ESC Heart Fail. 2021;8(1):222–237. doi:10.1002/ehf2.13144
5. Houssari M, Dumesnil A, Tardif V, et al. Lymphatic and immune cell cross-talk regulates cardiac recovery after experimental myocardial infarction. Arterioscler Thromb Vasc Biol. 2020;40(7):1722–1737. doi:10.1161/ATVBAHA.120.314370
6. Murphy A, Goldberg S. Mechanical Complications of Myocardial Infarction. Am J Med. 2022;135(12):1401–1409. doi:10.1016/j.amjmed.2022.08.017
7. Gong FF, Vaitenas I, Malaisrie SC, et al. Mechanical complications of acute myocardial infarction: a review. JAMA Cardiol. 2021;6(3):341–349. doi:10.1001/jamacardio.2020.3690
8. Strassheim D, Dempsey EC, Gerasimovskaya E, et al. Role of inflammatory cell subtypes in heart failure. J Immunol Res. 2019;2019(2164017):1–9. doi:10.1155/2019/2164017
9. Heinrichs M, Ashour D, Siegel J, et al. The healing myocardium mobilizes a distinct B-cell subset through a CXCL13-CXCR5-dependent mechanism. Cardiovasc Res. 2021;117(13):2664–2676. doi:10.1093/cvr/cvab181
10. Tang Y, Zeng X, Feng Y, et al. Corrigendum: association of systemic immune-inflammation index with short-term mortality of congestive heart failure: a retrospective cohort study. Front Cardiovasc Med. 2022;9:1116547.
11. Frampton JE. Empagliflozin: a review in symptomatic chronic heart failure. Drugs. 2022;82(16):1591–1602. doi:10.1007/s40265-022-01778-0
12. Zannad F, Ferreira JP, Pocock SJ, et al. SGLT2 inhibitors in patients with heart failure with reduced ejection fraction: a meta-analysis of the EMPEROR-Reduced and DAPA-HF trials. Lancet. 2020;396(10254):819–829. doi:10.1016/S0140-6736(20)31824-9
13. Anker SD, Butler J, Filippatos G, et al. Empagliflozin in heart failure with a preserved ejection fraction. N Engl J Med. 2021;385(16):1451–1461. doi:10.1056/NEJMoa2107038
14. Xu L, Nagata N, Nagashimada M, et al. SGLT2 inhibition by empagliflozin promotes fat utilization and browning and attenuates inflammation and insulin resistance by polarizing M2 macrophages in diet-induced obese mice. EBioMedicine. 2017;20:137–149.
15. Meng Z, Liu X, Li T, et al. The SGLT2 inhibitor empagliflozin negatively regulates IL-17/IL-23 axis-mediated inflammatory responses in T2DM with NAFLD via the AMPK/mTOR/autophagy pathway. Int Immunopharmacol. 2021;94(107492):107492. doi:10.1016/j.intimp.2021.107492
16. Kolijn D, Pabel S, Tian Y, et al. Empagliflozin improves endothelial and cardiomyocyte function in human heart failure with preserved ejection fraction via reduced pro-inflammatory-oxidative pathways and protein kinase Gα oxidation. Cardiovasc Res. 2021;117(2):495–507. doi:10.1093/cvr/cvaa123
17. Saeidian AH, Youssefian L, Vahidnezhad H, et al. Research techniques made simple: whole-transcriptome sequencing by RNA-Seq for diagnosis of monogenic disorders. J Invest Dermatol. 2020;140(6):1117–1126.e1111. doi:10.1016/j.jid.2020.02.032
18. Nakao M, Shimizu I, Katsuumi G, et al. Empagliflozin maintains capillarization and improves cardiac function in a murine model of left ventricular pressure overload. Sci Rep. 2021;11(1):18384. doi:10.1038/s41598-021-97787-2
19. Zhu Y, Chen X, Guo L, et al. Acute sleep deprivation increases inflammation and aggravates heart failure after myocardial infarction. J Sleep Res. 2022;31(6):e13679. doi:10.1111/jsr.13679
20. Sun D, Yang F. Metformin improves cardiac function in mice with heart failure after myocardial infarction by regulating mitochondrial energy metabolism. Biochem Biophys Res Commun. 2017;486(2):329–335. doi:10.1016/j.bbrc.2017.03.036
21. Emmons-Bell S, Johnson C, Roth G. Prevalence, incidence and survival of heart failure: a systematic review. Heart. 2022;108(17):1351–1360. doi:10.1136/heartjnl-2021-320131
22. Del Buono MG, Moroni F, Montone RA, et al. Ischemic cardiomyopathy and heart failure after acute myocardial infarction. Curr Cardiol Rep. 2022;24(10):1505–1515. doi:10.1007/s11886-022-01766-6
23. Andreadou I, Cabrera-Fuentes HA, Devaux Y, et al. Immune cells as targets for cardioprotection: new players and novel therapeutic opportunities. Cardiovasc Res. 2019;115(7):1117–1130. doi:10.1093/cvr/cvz050
24. Ong SB, Hernández-Reséndiz S, Crespo-Avilan GE, et al. Inflammation following acute myocardial infarction: multiple players, dynamic roles, and novel therapeutic opportunities. Pharmacol Ther. 2018;186:73–87.
25. Heidenreich PA, Bozkurt B, Aguilar D, et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: executive summary: a report of the American college of cardiology/American Heart Association joint committee on clinical practice guidelines. Circulation. 2022;145(18):e876–e894. doi:10.1161/CIR.0000000000001062
26. Kologrivova I, Shtatolkina M, Suslova T, et al. Cells of the immune system in cardiac remodeling: main players in resolution of inflammation and repair after myocardial infarction. Front Immunol. 2021;12(664457). doi:10.3389/fimmu.2021.664457
27. Komal S, Komal N, Mujtaba A, et al. Potential therapeutic strategies for myocardial infarction: the role of Toll-like receptors. Immunol Res. 2022;70(5):607–623. doi:10.1007/s12026-022-09290-z
28. Wang X, Li W, Zhang Y, et al. Calycosin as a Novel PI3K activator reduces inflammation and fibrosis in heart failure through AKT-IKK/STAT3 axis. Front Pharmacol. 2022;13:828061.
29. Yang Y, Xu C, Tang S, et al. Interleukin-9 aggravates isoproterenol-induced heart failure by activating signal transducer and activator of transcription 3 signalling. Can J Cardiol. 2020;36(11):1770–1781. doi:10.1016/j.cjca.2020.01.011
30. Szabo TM, Frigy A, Nagy EE. Targeting mediators of inflammation in heart failure: a short synthesis of experimental and clinical results. Int J Mol Sci. 2021;22(23):13053. doi:10.3390/ijms222313053
31. Morante-Palacios O, Fondelli F, Ballestar E, et al. Tolerogenic dendritic cells in autoimmunity and inflammatory diseases. Trends Immunol. 2021;42(1):59–75. doi:10.1016/j.it.2020.11.001
32. Choo EH, Lee JH, Park EH, et al. Infarcted myocardium-primed dendritic cells improve remodeling and cardiac function after myocardial infarction by modulating the regulatory T cell and macrophage polarization. Circulation. 2017;135(15):1444–1457. doi:10.1161/CIRCULATIONAHA.116.023106
33. Cao W, Chen J, Chen Y, et al. Advanced glycation end products promote heart failure through inducing the immune maturation of dendritic cells. Appl Biochem Biotechnol. 2014;172(8):4062–4077. doi:10.1007/s12010-014-0804-7
34. Silvestre-Roig C, Braster Q, Ortega-Gomez A, et al. Neutrophils as regulators of cardiovascular inflammation. Nat Rev Cardiol. 2020;17(6):327–340. doi:10.1038/s41569-019-0326-7
35. Tang X, Wang P, Zhang R, et al. KLF2 regulates neutrophil activation and thrombosis in cardiac hypertrophy and heart failure progression. J Clin Invest. 2022;132(3). doi:10.1172/JCI147191
36. Bermea K, Bhalodia A, Huff A, et al. The role of B cells in cardiomyopathy and heart failure. Curr Cardiol Rep. 2022;24(8):935–946. doi:10.1007/s11886-022-01722-4
37. Yang J, Li Y, Sun Z, et al. COL17A1 facilitates tumor growth and predicts poor prognosis in pancreatic cancer. Biochem Biophys Res Commun. 2022;632:1–9.
38. Maag JLV, Fisher OM, Levert-Mignon A, et al. Novel aberrations uncovered in barrett’s esophagus and esophageal adenocarcinoma using whole transcriptome sequencing. Mol Cancer Res. 2017;15(11):1558–1569. doi:10.1158/1541-7786.MCR-17-0332
39. Van den Bergh F, Eliason SL, Burmeister BT, et al. Collagen XVII (BP180) modulates keratinocyte expression of the proinflammatory chemokine, IL-8. Exp Dermatol. 2012;21(8):605–611. doi:10.1111/j.1600-0625.2012.01529.x
40. Lothong M, Sakares W, Rojsitthisak P, et al. Collagen XVII inhibits breast cancer cell proliferation and growth through deactivation of the AKT/mTOR signaling pathway. PLoS One. 2021;16(7):e0255179. doi:10.1371/journal.pone.0255179
41. Nikolov A, Popovski N. Extracellular matrix in heart disease: focus on circulating collagen type I and III derived peptides as biomarkers of myocardial fibrosis and their potential in the prognosis of heart failure: a concise review. Metabolites. 2022;12(4):297. doi:10.3390/metabo12040297
42. Papandreou C, Hernández-Alonso P, Bulló M, et al. High plasma glutamate and a low glutamine-to-glutamate ratio are associated with increased risk of heart failure but not atrial fibrillation in the prevención con dieta mediterránea (PREDIMED) Study. J Nutr. 2020;150(11):2882–2889. doi:10.1093/jn/nxaa273
43. Song J, Meng Y, Wang M, et al. Mangiferin activates Nrf2 to attenuate cardiac fibrosis via redistributing glutaminolysis-derived glutamate. Pharmacol Res. 2020;157(104845):104845. doi:10.1016/j.phrs.2020.104845
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.