Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 17
Identification of Dysregulated Mechanisms and Candidate Gene Markers in Chronic Obstructive Pulmonary Disease
Authors Lin J, Xue Y, Su W, Zhang Z, Wei Q, Huang T
Received 25 November 2021
Accepted for publication 27 February 2022
Published 4 March 2022 Volume 2022:17 Pages 475—487
DOI https://doi.org/10.2147/COPD.S349694
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Min Zhang
Jie Lin,1,2,* Yanlong Xue,1,2,* Wenyan Su,1,2,* Zan Zhang,1,2 Qiu Wei,1,2 Tianxia Huang1,2
1Department of Respiratory and Critical Care, The Fifth Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, 530022, People’s Republic of China; 2Department of Respiratory and Critical Care, The First People’s Hospital of Nanning, Nanning, Guangxi, 530022, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qiu Wei; Tianxia Huang, Department of Respiratory and Critical Care, The Fifth Affiliated Hospital of Guangxi Medical University, 89 Qixing Road, Nanning, Guangxi, 530022, People’s Republic of China, Tel +86 7712636163, Fax +86 7712617892, Email [email protected]; [email protected]
Purpose: This study aimed to identify candidate gene markers that may facilitate chronic obstructive pulmonary disease (COPD) diagnosis and treatment.
Methods: The GSE47460 and GSE151052 datasets were analyzed to identify differentially expressed mRNAs (DEmRs) between COPD patients and controls. DEmRs that were differentially expressed in the same direction in both datasets were analyzed for functional enrichment and for coexpression. Genes from the largest three modules were tested for their ability to diagnose COPD based on the area under the receiver operating characteristic curve (AUC). Genes with AUC > 0.7 in both datasets were used to perform regression based on the “least absolute shrinkage and selection operator” in order to identify feature genes. We also identified differentially expressed miRNAs (DEmiRs) between COPD patients and controls using the GSE38974 dataset, then constructed a regulatory network. We also examined associations between feature genes and immune cell infiltration in COPD, and we identified methylation markers of COPD using the GSE63704 dataset.
Results: A total of 1350 genes differentially regulated in the same direction in the GSE47460 and GSE151052 datasets were found. The genes were significantly enriched in immune-related biological functions. Of 186 modules identified using MEGENA, the largest were C1_ 6, C1_ 3, and C1_ 2. Of the 22 candidate genes screened based on AUC, 11 feature genes emerged from analysis of a subset of GSE47460 data, which we validated using another subset of GSE47460 data as well as the independent GSE151052 dataset. Feature genes correlated significantly with infiltration by immune cells. The feature genes GPC4 and RS1 were predicted to be regulated by miR-374a-3p. We identified 117 candidate methylation markers of COPD, including PRRG4.
Conclusion: The feature genes we identified may be potential diagnostic markers and therapeutic targets in COPD. These findings provide new leads for exploring disease mechanisms and targeted treatments.
Keywords: chronic obstructive pulmonary disease, bioinformatics analysis, miRNAs, immune response, feature genes
Introduction
According to estimates from the Global Burden of Disease (GBD) study, 523 million around the world had cardiovascular disease in 2019.1 Chronic obstructive pulmonary disease (COPD) is a major cause of global morbidity and mortality related to cardiovascular disease. COPD is caused by persistent, often progressive airflow limitation in the lungs, and it includes the conditions of chronic bronchitis and emphysema.2 More than 3 million people die from COPD every year worldwide,3 and COPD is expected to become the third leading cause of death globally by 2030.4 Despite the worldwide prevalence of this disease, it remains largely underdiagnosed and undertreated.
Many factors have been associated with COPD, including systemic and local inflammation, air pollution and smoking.5 However, the exact mechanism of COPD remains unclear. Clinically, this disorder is characterized by cough and chronic dyspnea resulting from airway stenosis. Accumulating evidence indicates that COPD is a systemic disease, and that its pathological manifestations are not limited to lung inflammation and airway remodeling6 In fact, COPD has a profound effect on cardiac function and gas exchange, and COPD patients are at 2–5 times greater risk of cardiovascular disease than the general population.7 Cardiovascular disease in any stage of COPD greatly increases the risk of death and hospitalization.8 Approximately 22–40% of COPD patients experience at least one moderate or severe exacerbation each year, and 9–16% experience more than one exacerbation per year.9
Dyspnea is the main symptom of COPD and the most frequent reason why COPD patients seek medical attention.10 At the onset of COPD, timely intervention to relieve symptoms and exacerbations can prevent an acute decline in lung function and progression of the condition to severe emphysema.11 Current treatments of COPD involve long-term inhalation therapy with bronchodilators, corticosteroids, or a combination of these agents.12 Combination therapy may be more effective than monotherapies for relieving COPD exacerbations, but the evidence is inconclusive.13 A challenge to effective treatment is that the nine approved drug classes for COPD maintenance therapy treat only the symptoms, rather than the underlying inflammation or progression.14
Elucidating the pathogenic pathways in COPD may help identify new treatments and strategies to prevent exacerbations. Toward that end, we applied bioinformatics to public databases to explore molecular mechanisms and feature genes potentially associated with COPD. We explored the potential functions of the feature genes in the disease, with a focus on immune cell infiltration as well as gene regulation by microRNAs (miRNAs) and DNA methylation. Understanding these regulatory mechanisms and identifying potential marker genes may lead to the development of new therapeutic strategies.
Materials and Methods
Data Collection
All data in this study were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The GSE47460 dataset15 included mRNA expression profiles from whole-lung homogenates of 144 COPD patients and 91 controls, obtained using the GPL14550 platform. The GSE151052 dataset16 included mRNA expression profiles from lung tissues of 77 COPD patients and 40 controls, obtained using the GPL17556 platform. The GSE38974 dataset17 included mRNA expression profiles from lung tissues of 23 COPD patients and 9 controls, obtained using the GPL4133 platform; as well as miRNA expression profiles from 19 COPD patients and 8 controls, obtained using the GPL7723 platform. The GSE76925 dataset18 included mRNA expression profiles from lung tissues of 111 COPD patients and 40 controls, obtained using the GPL10558 platform. The GSE63704 dataset19 included methylation profiles from lung tissues of 86 COPD patients and 26 controls, obtained using the GPL13534 platform.
Analysis of Expression and Methylation Differences Between COPD Patients and Controls
The limma package in R20 was used to search for differences in mRNA levels between COPD patients and controls in the GSE47460 and GSE151052 datasets, as well as differences in miRNA levels between COPD patients and controls in the GSE38974 dataset. The cutoff value for differential gene expression was P < 0.05. DEmRs and DEmiRs were defined as differentially expressed mRNAs or miRNAs, respectively. DEmRs that were differentially expressed (up- or downregulated) in the same direction in both the GSE47460 and GSE151052 datasets were defined as common genes.
The cAMP package in R was used to identify differences in gene methylation between COPD patients and controls in the GSE63704 dataset. Differentially methylated probes (DMPs) were defined as methylation sites that passed the significance threshold of P < 0.05. Only DMPs with ∆Beta values that varied between patients and controls in the opposite direction as common genes were retained in the analysis.
Analysis of Functional Enrichment
Common genes were analyzed for enrichment in Gene Ontology (GO) biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using the clusterProfiler package in R.21 Enrichment was considered significant at P < 0.05. Activated or inhibited enrichment was assessed using gene set variation analysis (GSVA) in R.22 Gene set enrichment analysis (GSEA) was also carried out using the fgsea package in R.
Construction of a Gene Coexpression Network
Common genes were analyzed using multiscale embedded gene coexpression network analysis (MEGENA) in R.23 This analysis generates embedded, multi-scale networks to uncover biologically meaningful genes, which are then assigned to modules of coexpressed genes.
Regression Based on the Least Absolute Shrinkage and Selection Operator (LASSO)
Genes in modules 2, 3, and 6 were assessed for their ability to differentiate COPD patients and controls based on the area under the receiver operating characteristic curve (AUC), as calculated using the pROC package in R.24 DEmRs that were present in both GSE47460 and GSE151052 datasets and that gave AUC > 0.7 were used to build a binomial LASSO regression model in the glmnet package in R.25 LASSO regression was performed to shrink the regression coefficients towards zero as λ increased. We then optimized λ in order to identify feature genes.
Immune Cell Infiltration
Levels of infiltration by different types of immune cells in lung tissues of COPD were evaluated using single‐sample GSEA as coded in the GSVA package in R. The analysis was performed on data from datasets GSE151052, GSE38974, GSE47460, and GSE76925. Differences in infiltration levels between COPD patients and controls were identified using the limma package. We also evaluated potential correlations between feature gene expression and levels of immune cell infiltration using Pearson correlation analysis. Results associated with P < 0.05 were considered significant. Proportions of immune cells in lung tissues of COPD patients were analyzed using CIBERSORT (https://cibersort.stanford.edu/), after excluding immune cell types associated with “0”.
Target Prediction
Downstream genes possibly regulated by DEmiRs were predicted using Targetscan (http://www.targetscan.org/vert_72/).
Statistical Analysis
All statistical analyses were performed using R (version 3.9.1) and related packages within the R environment. Results associated with P < 0.05 were considered statistically significant.
Results
DEmRs in COPD
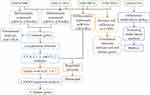
The design of this study is shown in Figure 1. We identified 6544 DEmRs between COPD patients and controls in the GSE47460 dataset (Figure 2A, Table S1) and 10,693 DEmRs in the GSE151052 dataset (Figure 2B, Table S2), based on a definition of differential expression as fold change > 0. A total of 487 DEmRs were upregulated in both datasets (Figure 2C), while 863 were downregulated in both (Figure 2D). These DEmRs were defined as common genes, which may be linked to COPD disease.
Biological Functions of Common Genes
To predict the biological functions in which common genes may be involved, we performed enrichment analysis. Enrichment analysis implicated the common genes in the GO biological processes of T cell activation, regulation of leukocyte activation, and lymphocyte differentiation (Figure 3A). The common genes were also implicated in the KEGG signaling pathways of leukocyte transendothelial migration, primary immunodeficiency, and asthma (Figure 3B). GSEA of common genes suggested that COPD was associated with activation of the intestinal immune network involved in IgA production, with activation of asthma pathways, and with inhibition of axon guidance and tight junctions (Figure 3C). These results suggest that common genes are involved in biological processes related to immune and inflammatory responses.
Construction of a Gene Coexpression Network
To further identify aberrantly expressed genes with important roles in COPD, we performed MEGENA on common genes (Figure 4A), which defined 186 modules containing a total of 1345 genes. The three largest modules were C1_ 6, with 270 genes; C1_3, 269 genes; and C1_2, 259 genes (Figure 4B). Genes in these three modules that differentiated COPD patients from controls with an AUC > 0.7 in both the GSE47460 and GSE151052 datasets (Figure 4C) were defined as candidate genes. We obtained 22 candidate genes (Figure 4D), which may be useful in diagnosing COPD.
Identification of Feature Genes in COPD
We used LASSO regression to select which of the 22 candidate genes were most likely to be important in COPD, based on the GSE47460 dataset. Based on an optimized λ of 0.02161155, we obtained 11 feature genes with non-zero coefficients (Figure 5A and B): AQP1, CLEC4A, GPC4, GPR39, PDCL3, PGM1, PRRG4, ROR1, RS1, SEC14L4, and USP13. After randomly dividing up the COPD patient samples in the GSE47460 dataset into a training set (75%) and validation set (25%), we evaluated the diagnostic performance of the feature genes against the training set based on AUC (Figure 5C). The AUC for all feature genes together was 0.885 against the training set and 0.870 against the validation set (Figure 5D). The AUC was even higher (0.918) against independent external validation data, the GSE151052 dataset (Figure 5E).
Immune Cell Infiltration in COPD
Since common genes were enriched in functions related to immune responses, we hypothesized that immune cell dysfunction might play a key role in COPD. Indeed, we found that infiltration by Th1 cells, follicular helper T cell (TFH), CD8+ T cells, and B cells was significantly greater in COPD patients than in controls (Figure 6A). In fact, these four immune cell types correlated positively with one another (Figure 6B). Feature genes correlated significantly with levels of immune cell infiltration in COPD patients (Figure 6C), with macrophages as the most abundant infiltrating cells (Figure 6D).
Regulation of Feature Genes by miRNAs
To identify how miRNAs may regulate feature genes, we started from 223 DEmiRs between COPD patients and controls in the GSE38974 dataset (Figure 7A). Then we predicted the downstream mRNAs targeted by the following six miRNAs with the largest log | (fold change) |: hsa-miR-1274a, hsa-miR-105-5p, hsa-miR-374a-3p, hsa-miR-422a-3p, hsa-miR-937-5p, and hsa-miR-923. Of the predicted mRNA targets, 162 were present among DEmRs modules C1_6, C1_3, and C1_2. These 162 downstream target genes were enriched in divalent inorganic cation homeostasis, cellular response to nitrogen compounds, as well as glutamate and glutamine metabolism (Figure 7B). Two of the genes, GPC4 and RS1, were predicted to be regulated by miR-374a-3p (Figure 7C). The AUC for miR-374a-3p was 0.89, suggesting the ability to diagnose COPD (Figure 7D). Finally, we generated a map of DEmiRs regulating DEmRs, which were involved in cell junction assembly, positive regulation of the MAPK cascade, and VEGFA−VEGFR2 signaling (Figure 7E).
Aberrant Gene Methylation in COPD
In the GSE63704 dataset, we identified 6495 DMPs between COPD and control samples, involving 4296 genes (Figure 8A). After retaining only DMPs that were increased or decreased between patients and controls in the opposite direction as common genes, we were left with 117 potential methylation markers (Figure 8B). Among them, we found that the feature gene PRRG4 was aberrantly methylated in COPD.
Discussion
COPD is a common respiratory disease that seriously threatens human health and well-being, and it is an important public health problem.26 Current treatments for COPD aim to improve symptoms and prevent exacerbations, but none is disease-modifying.27 To provide the knowledge needed to improve therapy options, we compared gene expression profiles between COPD patients and healthy controls in order to identify coexpressed differentially expressed genes. MEGENA is an innovative method for analyzing coexpression networks that has advantages over weighted gene co-expression network analysis (WGCNA), facilitating efficient construction of large-scale coexpression plane filter networks while preserving gene interactions.23 Our MEGENA identified 186 functional modules, the largest of which were C1_ 6, C1_ 3, and C1_ 2. Next we screened the differentially expressed genes using AUCs and LASSO regression to identify 11 feature genes, which we validated using an internal dataset (GSE47460) and external dataset (GSE151052). Feature genes have been widely used in multi-marker profiling studies.28–30 Our feature genes in COPD showed high AUCs, suggesting their potential for clinical applications.
We found that many of the differentially expressed genes in COPD were associated with various immune responses. An increasing number of immune cell types have been associated with risk of COPD and prognosis of affected individuals.31,32 In particular, COPD has been linked to elevated numbers of neutrophils, B cells, as well as CD4+ and CD8+ T lymphocytes in the lungs.33 During adaptive immune responses in COPD patients, activated T cells can promote abnormal inflammatory responses and aggravate airway damage.32 Increased B cell counts in COPD patients have been associated with elevated IgA synthesis, which impairs mucosal immunity and may contribute to disease progression.34 Consistent with this, our GSEA found that the intestinal immune network involved in IgA production was activated in COPD patients. Genetic alterations in leukocyte transendothelial migration pathways in smokers and COPD patients have been strongly associated with T cell levels and airway obstruction,35,36 which our enrichment analyses support. In addition, asthma has been associated with the frequency and severity of COPD exacerbations,37 which our analysis of DEmR enrichment supports.
Feature genes of COPD that we identified here have previously been linked to the disease, suggesting that our bioinformatics analysis is reliable. AQP1 is significantly up-regulated in COPD and has proven to be an effective therapeutic target.38,39 Expression of CLEC4A is influenced by smoking,40 which strongly correlates with risk of COPD and airflow obstruction.41,42 GPC4, which remodels the extracellular matrix, is closely related to COPD disease regions.43,44 GPR39 helps activate pro-inflammatory signaling pathways,45 which may contribute to pathological inflammation in COPD. PDCL3 regulates expression of VEGF receptor 2, and it may promote pathological angiogenesis.46,47 PGM1 has been linked to lung disease through multiple metabolic pathways.48,49 PRRG4 has been shown to be differentially expressed in COPD patients,50 and the present study not only confirms that finding but extends it by showing aberrant methylation of the gene. Therefore, the altered expression of PRRG4 in COPD may reflect altered gene methylation. ROR1, after binding to Wnt ligands, triggers non-canonical signaling cascades that increase the level of calcium or decrease the level of cGMP within the cell, and these changes are closely associated with COPD onset and progression.51 SEC14L4 is known to be differentially expressed in COPD.52 USP13, which inhibits autophagy, may be involved in COPD pathogenesis.53,54
Among the 11 feature genes, RS1 has not previously been linked to COPD, and our study justifies further exploration of this potential link. In addition, our results suggest that the gene, together with GPC4, is regulated by miR-374a-3p. This miRNA has been shown to regulate inflammatory responses.55,56 Further work should examine whether miR-374a-3p is involved in COPD via its ability to regulate the expression of RS1 and GPC4.
Our findings should be interpreted with caution in light of several limitations. We were limited to data available in public databases, and we did not validate key results using independent clinical samples or biochemical experiments. In addition, relevant clinical and follow-up data were unavailable for many of the samples that we analyzed, preventing us from examining potential relationships between feature genes and comorbidities or prognosis. Further study with larger samples and detailed follow-up should verify and extend the clinical utility of the feature genes that we identified here. Experimental studies in vivo and in vitro should explore how feature genes in COPD are regulated.
Conclusion
Many genes that are abnormally expressed in COPD are involved in immune responses, and we identified several feature genes that may be potential markers and therapeutic targets in COPD. In this way, our bioinformatics study generates numerous leads to guide future research into the disease and its treatment.
Abbreviations
AUC, area under the receiver operating characteristic curve; COPD, chronic obstructive pulmonary disease; DEmiR, differentially expressed miRNA; DEmR, differentially expressed mRNA; DMP, differentially methylated probe; GBD; Global Burden of Disease; GEO, Gene Expression Omnibus; GO, Gene Ontology; GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; LASSO, least absolute shrinkage and selection operator; MEGENA, multiscale embedded gene coexpression network analysis; miRNA, microRNA.
Data Sharing Statement
The raw data, analyses and codes in this study can be obtained from the corresponding author upon reasonable request.
Ethics Approval and Informed Consent
Such approval or consent was not required by the Ethics Committee of the Fifth Affiliated Hospital of Guangxi Medical University because this study was based entirely on publicly available, freely downloadable data, for which the original submitters were required to obtain relevant ethics approval and consent.
Acknowledgments
This study was supported by the Project of Qingxiu District of Nanning Scientific Research and Technology Development Plan (2019038), the Project of Nanning Scientific Research and Technology Development Plan (20183040-4 and ZC20213005), Guangxi medical and health key discipline construction project (Department of Respiratory and Critical Care, The First People’s Hospital of Nanning).
Disclosure
The authors have no potential conflicts of interest to declare in this work.
References
1. Roth GA, Mensah GA, Johnson CO, et al.; GBD-NHLBI-JACC Global Burden of Cardiovascular Diseases Writing Group. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol. 2020;76(25):2982–3021. doi:10.1016/j.jacc.2020.11.010
2. Karnati S, Seimetz M, Kleefeldt F, et al. Chronic obstructive pulmonary disease and the cardiovascular system: vascular repair and regeneration as a therapeutic target. Front Cardiovasc Med. 2021;8:649512. doi:10.3389/fcvm.2021.649512
3. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389(10082):1931–1940. doi:10.1016/S0140-6736(17)31222-9
4. Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. doi:10.1371/journal.pmed.0030442
5. Yao Y, Zhou J, Diao X, Wang S. Association between tumor necrosis factor-alpha and chronic obstructive pulmonary disease: a systematic review and meta-analysis. Ther Adv Respir Dis. 2019;13:1753466619866096. doi:10.1177/1753466619866096
6. Chan SMH, Selemidis S, Bozinovski S, Vlahos R. Pathobiological mechanisms underlying metabolic syndrome (MetS) in chronic obstructive pulmonary disease (COPD): clinical significance and therapeutic strategies. Pharmacol Ther. 2019;198:160–188. doi:10.1016/j.pharmthera.2019.02.013
7. Chen W, Thomas J, Sadatsafavi M, FitzGerald JM. Risk of cardiovascular comorbidity in patients with chronic obstructive pulmonary disease: a systematic review and meta-analysis. Lancet Respir Med. 2015;3(8):631–639. doi:10.1016/S2213-2600(15)00241-6
8. Franssen FM, Rochester CL. Comorbidities in patients with COPD and pulmonary rehabilitation: do they matter? Eur Respir Rev. 2014;23(131):131–141. doi:10.1183/09059180.00007613
9. Mathioudakis AG, Janssens W, Sivapalan P, et al. Acute exacerbations of chronic obstructive pulmonary disease: in search of diagnostic biomarkers and treatable traits. Thorax. 2020;75(6):520–527. doi:10.1136/thoraxjnl-2019-214484
10. Sandelowsky H, Weinreich UM, Aarli BB, et al. COPD - do the right thing. BMC Fam Pract. 2021;22(1):244. doi:10.1186/s12875-021-01583-w
11. Jung T, Vij N. Early diagnosis and real-time monitoring of regional lung function changes to prevent chronic obstructive pulmonary disease progression to severe emphysema. J Clin Med. 2021;10(24):5811. doi:10.3390/jcm10245811
12. Yang YL, Xiang ZJ, Yang JH, Wang WJ, Xu ZC, Xiang RL. Association of beta-blocker use with survival and pulmonary function in patients with chronic obstructive pulmonary and cardiovascular disease: a systematic review and meta-analysis. Eur Heart J. 2020;41(46):4415–4422. doi:10.1093/eurheartj/ehaa793
13. Oba Y, Keeney E, Ghatehorde N, Dias S. Dual combination therapy versus long-acting bronchodilators alone for chronic obstructive pulmonary disease (COPD): a systematic review and network meta-analysis. Cochrane Database Syst Rev. 2018;12:CD012620. doi:10.1002/14651858.CD012620.pub2
14. van Haarst A, McGarvey L, Paglialunga S. Review of drug development guidance to treat chronic obstructive pulmonary disease: US and EU perspectives. Clin Pharmacol Ther. 2019;106(6):1222–1235. doi:10.1002/cpt.1540
15. Kim S, Herazo-Maya JD, Kang DD, et al. Integrative phenotyping framework (iPF): integrative clustering of multiple omics data identifies novel lung disease subphenotypes. BMC Genomics. 2015;16:924. doi:10.1186/s12864-015-2170-4
16. Su X, Chen J, Lin X, et al. FERMT3 mediates cigarette smoke-induced epithelial-mesenchymal transition through Wnt/beta-catenin signaling. Respir Res. 2021;22(1):286. doi:10.1186/s12931-021-01881-y
17. Ezzie ME, Crawford M, Cho JH, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67(2):122–131. doi:10.1136/thoraxjnl-2011-200089
18. Morrow JD, Zhou X, Lao T, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep. 2017;7:44232. doi:10.1038/srep44232
19. Wielscher M, Vierlinger K, Kegler U, Ziesche R, Gsur A, Weinhausel A. Diagnostic performance of plasma DNA methylation profiles in lung cancer, pulmonary fibrosis and COPD. EBioMedicine. 2015;2(8):929–936. doi:10.1016/j.ebiom.2015.06.025
20. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
21. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
22. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
23. Song WM, Zhang B. Multiscale embedded gene co-expression network analysis. PLoS Comput Biol. 2015;11(11):e1004574. doi:10.1371/journal.pcbi.1004574
24. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi:10.1186/1471-2105-12-77
25. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. doi:10.18637/jss.v033.i01
26. Quan Z, Yan G, Wang Z, et al. Current status and preventive strategies of chronic obstructive pulmonary disease in China: a literature review. J Thorac Dis. 2021;13(6):3865–3877. doi:10.21037/jtd-20-2051
27. Brandsma CA, Van den Berge M, Hackett TL, Brusselle G, Timens W. Recent advances in chronic obstructive pulmonary disease pathogenesis: from disease mechanisms to precision medicine. J Pathol. 2020;250(5):624–635. doi:10.1002/path.5364
28. Tian S, Meng G, Zhang W. A six-mRNA prognostic model to predict survival in head and neck squamous cell carcinoma. Cancer Manag Res. 2019;11:131–142. doi:10.2147/CMAR.S185875
29. Siow ZR, De Boer RH, Lindeman GJ, Mann GB. Spotlight on the utility of the Oncotype DX® breast cancer assay. Int J Womens Health. 2018;10:89–100. doi:10.2147/IJWH.S124520
30. Wang SY, Dang W, Richman I, Mougalian SS, Evans SB, Gross CP. Cost-effectiveness analyses of the 21-gene assay in breast cancer: systematic review and critical appraisal. J Clin Oncol. 2018;36(16):1619–1627. doi:10.1200/JCO.2017.76.5941
31. Trivedi A, Khan MA, Bade G, Talwar A. Orchestration of neutrophil extracellular traps (nets), a unique innate immune function during Chronic Obstructive Pulmonary Disease (COPD) development. Biomedicines. 2021;9(1):53. doi:10.3390/biomedicines9010053
32. Schivo M, Albertson TE, Haczku A, et al. Paradigms in chronic obstructive pulmonary disease: phenotypes, immunobiology, and therapy with a focus on vascular disease. J Investig Med. 2017;65(6):953–963. doi:10.1136/jim-2016-000358
33. Szalontai K, Gemes N, Furak J, et al. Chronic obstructive pulmonary disease: epidemiology, biomarkers, and paving the way to lung cancer. J Clin Med. 2021;10(13):2889. doi:10.3390/jcm10132889
34. Polverino F, Seys LJ, Bracke KR, Owen CA. B cells in chronic obstructive pulmonary disease: moving to center stage. Am J Physiol Lung Cell Mol Physiol. 2016;311(4):L687–L695. doi:10.1152/ajplung.00304.2016
35. Yang M, Kohler M, Heyder T, et al. Long-term smoking alters abundance of over half of the proteome in bronchoalveolar lavage cell in smokers with normal spirometry, with effects on molecular pathways associated with COPD. Respir Res. 2018;19(1):40. doi:10.1186/s12931-017-0695-6
36. Su YC, Jalalvand F, Thegerstrom J, Riesbeck K. The interplay between immune response and bacterial infection in COPD: focus upon non-typeable Haemophilus influenzae. Front Immunol. 2018;9:2530. doi:10.3389/fimmu.2018.02530
37. Wang C, Zhou J, Wang J, et al. Progress in the mechanism and targeted drug therapy for COPD. Signal Transduct Target Ther. 2020;5(1):248. doi:10.1038/s41392-020-00345-x
38. Wu X, Sun X, Chen C, Bai C, Wang X. Dynamic gene expressions of peripheral blood mononuclear cells in patients with acute exacerbation of chronic obstructive pulmonary disease: a preliminary study. Crit Care. 2014;18(6):508. doi:10.1186/s13054-014-0508-y
39. Zhen G, Upur H, Jing W, et al. Effect of abnormal Savda Munziq, a traditional Uighur herbal medicine, on pulmonary function and aquaporins of COPD rat model with abnormal Savda syndrome. Evid Based Complement Alternat Med. 2017;2017:7176263. doi:10.1155/2017/7176263
40. Carolan BJ, Harvey BG, De BP, Vanni H, Crystal RG. Decreased expression of intelectin 1 in the human airway epithelium of smokers compared to nonsmokers. J Immunol. 2008;181(8):5760–5767. doi:10.4049/jimmunol.181.8.5760
41. Vogelmeier CF, Criner GJ, Martinez FJ, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. 2017;195(5):557–582. doi:10.1164/rccm.201701-0218PP
42. Hatipoglu U. Chronic obstructive pulmonary disease: more than meets the eye. Ann Thorac Med. 2018;13(1):1–6. doi:10.4103/atm.ATM_193_17
43. Adams TS, Schupp JC, Poli S, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv. 2020;6(28):eaba1983. doi:10.1126/sciadv.aba1983
44. Sharma A, Kitsak M, Cho MH, et al. Integration of molecular interactome and targeted interaction analysis to identify a COPD disease network module. Sci Rep. 2018;8(1):14439. doi:10.1038/s41598-018-32173-z
45. Voelkl J, Egli-Spichtig D, Alesutan I, Wagner CA. Inflammation: a putative link between phosphate metabolism and cardiovascular disease. Clin Sci. 2021;135(1):201–227.
46. Srinivasan S, Meyer RD, Lugo R, Rahimi N. Identification of PDCL3 as a novel chaperone protein involved in the generation of functional VEGF receptor 2. J Biol Chem. 2013;288(32):23171–23181. doi:10.1074/jbc.M113.473173
47. Srinivasan S, Chitalia V, Meyer RD, et al. Hypoxia-induced expression of phosducin-like 3 regulates expression of VEGFR-2 and promotes angiogenesis. Angiogenesis. 2015;18(4):449–462. doi:10.1007/s10456-015-9468-3
48. Bhargava M, Viken KJ, Barkes B, et al. Novel protein pathways in development and progression of pulmonary sarcoidosis. Sci Rep. 2020;10(1):13282. doi:10.1038/s41598-020-69281-8
49. Arakelyan A, Nersisyan L, Petrek M, Loffler-Wirth H, Binder H. Cartography of pathway signal perturbations identifies distinct molecular pathomechanisms in malignant and chronic lung diseases. Front Genet. 2016;7:79. doi:10.3389/fgene.2016.00079
50. Lin YZ, Zhong XN, Chen X, Liang Y, Zhang H, Zhu DL. Roundabout signaling pathway involved in the pathogenesis of COPD by integrative bioinformatics analysis. Int J Chron Obstruct Pulmon Dis. 2019;14:2145–2162. doi:10.2147/COPD.S216050
51. Shi J, Li F, Luo M, Wei J, Liu X. Distinct roles of Wnt/beta-catenin signaling in the pathogenesis of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Mediators Inflamm. 2017;2017:3520581. doi:10.1155/2017/3520581
52. Cruz T, Lopez-Giraldo A, Noell G, et al. Multi-level immune response network in mild-moderate Chronic Obstructive Pulmonary Disease (COPD). Respir Res. 2019;20(1):152. doi:10.1186/s12931-019-1105-z
53. Pesce E, Sondo E, Ferrera L, et al. The autophagy inhibitor spautin-1 antagonizes rescue of mutant CFTR through an autophagy-independent and USP13-mediated mechanism. Front Pharmacol. 2018;9:1464. doi:10.3389/fphar.2018.01464
54. Ornatowski W, Lu Q, Yegambaram M, et al. Complex interplay between autophagy and oxidative stress in the development of pulmonary disease. Redox Biol. 2020;36:101679. doi:10.1016/j.redox.2020.101679
55. Wang M, Wei J, Shang F, Zang K, Zhang P. Down-regulation of lncRNA SNHG5 relieves sepsis-induced acute kidney injury by regulating the miR-374a-3p/TLR4/NF-kappaB pathway. J Biochem. 2021;169(5):575–583. doi:10.1093/jb/mvab008
56. Shi FL, Ren LX. Up-regulated miR-374a-3p relieves lipopolysaccharides induced injury in CHON-001 cells via regulating Wingless-type MMTV integration site family member 5B. Mol Cell Probes. 2020;51:101541. doi:10.1016/j.mcp.2020.101541
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.