Back to Journals » Cancer Management and Research » Volume 11
Identification of chromosomal abnormalities and genomic features in near-triploidy/tetraploidy-acute leukemia by fluorescence in situ hybridization
Authors Yang R, Jiang M , Zhao J, Chen H, Gong J, You Y, Song L, Li Z, Li Q
Received 28 September 2018
Accepted for publication 27 December 2018
Published 15 February 2019 Volume 2019:11 Pages 1559—1567
DOI https://doi.org/10.2147/CMAR.S189025
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Beicheng Sun
Ruqing Yang,1 Minghua Jiang,2 Junzhao Zhao,1 Hui Chen,2 Jian Gong,2 Yaying You,3 Laiyue Song,3 Zhen Li,4 Qian Li2
1Reproductive Medicine Center, Department of Gynaecology and Obstetrics, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou, Zhejiang 325000, China; 2Department of Clinical Laboratory, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou, Zhejiang 325000, China; 3The Second Clinical Medical College, Wenzhou Medical University, Wenzhou, Zhejiang 325000, China; 4Department of Traditional Chinese Medicine, The Third Affiliated Hospital of Guangzhou Medical University, Key Laboratory for Major Obstetric Diseases of Guangdong Province, Guangzhou, Guangdong 510150, China
Background: Near-triploidy/tetraploidy is rarely found in acute leukemia. Only limited data are available to characterize this condition, and it remains largely unknown.
Patients and methods: In our study, we performed karyotype analysis on 1,031 patients diagnosed with acute leukemia from 2006 to 2018. A total of 10 patients of near-triploidy/tetraploidy karyotype were enrolled. Two cases of near-triploidy (66–79 chromosomes) and eight cases of near-tetraploidy (84–100 chromosomes) were identified. Bone marrow samples of these 10 patients were analyzed by fluorescence in situ hybridization with 19 commercially available probes that detected a small portion of gene alterations and large regions of chromosome amplifications.
Results: Of the six patients with acute myelocytic leukemia, we detected three cases of double t(8;21)(q22;q22) that have not been previously reported, and one of them demonstrated ins(21;8)(q22;q24q22). We also describe a novel pediatric case carrying double t(15;17)(q22;q21) and receiving targeted treatment with all-trans retinoic acid therapy. To date, this case has responded well to the regimen and has shown continuous complete remission. All patients received chemotherapy. One of them received allogeneic hematopoietic stem cell transplant (HSCT) and survived for 22 months. Eight of the 10 patients died, and the median overall survival was 11 months.
Conclusion: Using fluorescence in situ hybridization, we identified the distinct complex karyotype of near-triploidy/tetraploidy and provided further prognostic information. Tetraploidy acute promyelocytic leukemia had favorable prognosis; thus, HSCT was not necessary. The case of insertion t(21;8)(q22;q24q22) in tetraploidy responded poorly to chemotherapy and achieved molecular remission with difficultly. Data from patients in this group indicated that near-triploidy/tetraploidy acute leukemia has poor prognosis and new therapy is urgently needed.
Keywords: near-triploidy, tetraploidy karyotype, acute leukemia, fluorescence in situ hybridization, gene
Introduction
Near-triploidy/tetraploidy is a rare cytogenetic abnormality in acute leukemia (AL), often involving numerical and structural abnormalities.1–3 The near-triploidy karyotype often originates from a duplication of low hypodiploid karyotype in acute lymphoblastic leukemia (ALL) and is associated with poor prognosis.4 However, near-triploidy karyotype is frequently misinterpreted as high hyperdiploid karyotype with favorable prognosis.5,6 So far, only limited data are available because of the low incidence of this ALL subset, affecting only 1% of B lineage ALL.7 However, near-tetraploidy in acute myelocytic leukemia (AML) is usually divided into two categories: primary and secondary.8 Primary near-tetraploidy AML originates from pluripotent myeloid progenitor cells, and most metaphase cells demonstrate near-tetraploidy karyotypes. Secondary near-tetraploidy AML originates from further differentiated myeloid precursor cells and is often associated with a duplication of abnormal chromosomes.9 Xue et al reported six cases of secondary near-tetraploidy AML. This rare secondary near-tetraploidy was characterized by double t(8;21) leukemia, which might be associated with poor prognosis.10,11
Unfortunately, the karyotype of this entity is complex and the precise numerical and structural abnormalities are difficult to distinguish by conventional karyotype analysis. Thus, in most routine clinical diagnostic laboratories, fluorescence in situ hybridization (FISH) is the gold standard to overcome this limitation.12,13
To better characterize this entity, we analyzed 10 patients with near-triploidy/tetraploidy-AL by FISH. Through inter/metaphase FISH, we identified a complex karyotype with multiple chromosomal abnormalities and marker chromosomes. In addition, FISH detected a small percentage of genomic variations. Overall, FISH can help identify the mechanisms involved in forming such karyotypes and offer information on the prognostic impact of gene alterations and chromosomal abnormalities.
Patients and methods
Patients
We searched the database of the clinical cytogenetics laboratory at the Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University during the years 2006 and 2018. A total of 10 cases with near-triploidy/tetraploidy-AL were enrolled. We reviewed electronic records of patients’ data, including performance status, treatment regimen, response to induction therapy, and survival. The ethics committee of the Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University had approved our data collection and sample analysis. All patients completed the informed written consent (informed written consent was completed by a parent or guardian in regards to patients under the age of 18 years) in accordance with the Declaration of Helsinki.
Morphology and flow cytometric immunophenotyping
Bone marrow (BM) aspirate smears were reviewed in all cases. Cases were classified according to French–American–British criteria. Cell surface antigens were detected by flow cytometry (FACSC antoTM II; BD, Franklin Lakes, NJ, USA) and further analyzed by extended panels designed to characterize AL according to the methods described previously.14–16
Conventional karyotyping
BM cells were cultivated for 24–48 hours without mitogen stimulation and harvested for chromosomal examination in a standard way. At least 20 metaphases R-banded by the Giemsa stain were examined, and the International System for Human Cytogenetic Nomenclature (2009) was used to describe chromosomal abnormalities.
Molecular genetic analysis
RT-PCR analysis
Total RNA was extracted from the patients’ BM cells at the time of diagnosis and relapse. RT-PCR analyses were performed following the manufacturer’s instructions. For PML–RARA fusion gene, RT-PCR was performed to detect long and short PML–RARA chimeric transcripts by using total RNA essentially, as described previously.17 For AML–ETO, the primer sequences used was reported elsewhere.18,19
Fluorescence in situ hybridization
We used the 19 commercially available probes to detect 10 cases by inter/meta FISH. Interphase signals were evaluated in 200 nuclei of cells. Images were captured by a Nikon 80-A1 fluorescent microscope and analyzed with image analysis software AI.
Statistical analysis
All data were analyzed using the SPSS for Windows (ver. 16.0; SPSS Inc.). Overall survival (OS) was measured from the date of diagnosis until the last follow-up or death. P≤0.05 was considered to be statistically significant.
Results
Patients
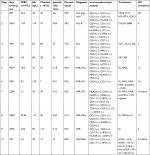
Between 2006 and 2018, a total of 1,031 patients were diagnosed with AL, but only 10 patients (3 females and 7 males) were enrolled in our project (10/1,031). The patients’ baseline demographic and clinical features are summarized in Table 1. Age ranged from 6 to 88 years, with a median age of 38.3 years. Eight (80%) were adults and two (20%) were children (age <18 years old). Six patients were classified as AML and the remaining four were ALL. Detailed case descriptions and analysis can be found in Tables 1 and 2; Figures 1-4; and Figures S1–S8.
  | Table 2 Karyotypes Notes: aAt the first diagnosis of the disease. bAt the first relapse. cContinuous complete remission. |
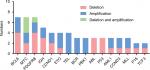  | Figure 4 Genes change in near-triploid/tetraploid-AL in 10 cases. Abbreviation: AL, acute leukemia. |
Laboratory, morphologic, and immunophenotypic findings
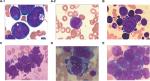
All patients’ laboratory findings are reported in Table 1. The median percentage of BM blasts was 69.5% (range, 40%–95.5%). BM aspirates showed active proliferation of leukemic cells and all cases showed large blasts, though some also had admixed blasts (Figure 1A-1, B, D, E). Auer rods and cytoplasmic granules could easily be seen in six AML cases (Figure 1A-2, C).
In AML, leukemic blasts frequently expressed myeloid-associated markers CD13 (5/6), CD33 (5/6), and CD117 (5/6).20,21 Case 7, with AML-M2, expressed not only classical myeloid-associated markers, but also CD19 and CD38 lymphoid-associated markers and CD56 antigen. Case 6 was diagnosed with gastric lymphoma, and immunohistochemistry analysis of a biopsy specimen found positivity for CD20, CD45RO, BCL-2, and LCA. About 10 years later, the patient was diagnosed with AML in diploid cells and expressed CD34 and CD117. Upon relapse after 9 months, the immunological analysis of the patient was not acquired. This phenomenon revealed that an AML immunophenotype might be accompanied by nontypical myeloid expression. This condition was also discovered in ALL. Case 3 showed typical myeloid-associated markers of CD13, CD15, and CD33 without any lymphoid-associated markers. Case 4 expressed CD16 and CD56. These findings suggest that immunophenotypic heterogeneity exists in near-triploidy/tetraploidy-AL.9,22
Cytogenetics
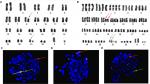
Karyotypes of all 10 patients are listed in Table 2. The chromosome numbers in these cases were within 70–97. A total of three cases demonstrated a coexisting diploid karyotype and three cases presented a coexisting high hypodiploidy (42–45 chromosomes)/low hyperdiploidy (47–49 chromosomes) karyotype. In this cohort, numerical alterations were seen in eight cases, structural alterations in nine cases, and one case (Case 1) showed a non-complex karyotype (Figure S8). Two cases with near-triploidy karyotype were both diagnosed with ALL (Case 2 and Case 4), and the remaining eight were near-tetraploidy karyotype. Three cases had lost chromosome 7, and one case had deletion of 7q. Thus, four cases had abnormal chromosome 7 (4/10; Figure 3A; Figures S1A, S2, S3A, B, D). Furthermore, two of four cases had concomitantly lost chromosome 13 (2/4) and one of four cases had an extra chromosome 22×2 (Figure 3A; Figures S1A, S3C). Case 3 had a tetraploidy karyotype both at diagnosis and relapse, as well as the amplification of 1q and deletion of 8q, respectively (Figure S4A, B). In Case 6, diploid clones were detected at first diagnosis, but near-tetraploidy clones were found at relapse, both accompanied by typical t(8;21). Moreover, the karyotype of t(8;21) was also observed in Case 5 and Case 7 (Figure 2A, B; Figure S5). The classical t(15;17) was found in Case 10 (Figure S6A).
Results of inter/metaphase FISH analysis
Inter/metaphase FISH results were obtained in all 10 cases. The details of 19 probes are listed in Table S1.
Translocation and isodicentric findings
We used GLP RUNX1–RUNX1T1 probes to detect clonal aberrations.23 In AML, a double t(8;21)(q22;q22) translocation occurred in three patients (30%) by karyotype analysis, but classical t(8;21) was detected in two cases by FISH (Figures S2 and S5). One insertion translocation of ins(21;8)(q22;q24q22) occurred in Case 7, which was further confirmed by GLP RUNX1–RUNX1T1, ETV6–RUNX1, and MYC probes (Figure 2C–E). Case 10 was a child with acute promyelocytic leukemia (APL), having classical double t(15,17)(q22;q21) translocation by karyotype analysis and FISH (Figure S6A, B). RT-PCR analysis again revealed the S-type isoform PML–RARA fusion transcript in APL cells. After chemotherapy, the patient presented a normal karyotype of 46, XY, N [20], but minimal residual disease continued to be positive for 4 months, followed by maintained continuous complete remission (CCR). Case 9 had isodicentric 11q chromosomes idic (11q), as confirmed by a GLP MLL (KMT2A) probe (located at 11q23) and an IGH–CCND1 (located at 14q32/11q13) probe, with the loss chromosomes of 5, 7, and 13 (Figure 3B; Table S2). Case 4 had isodicentric 3q chromosomes idic (3q) confirmed by a GLP BCL6 probe (Figure S7A; Table S2). Case 2 showed 22p+×2 confirmed by using a GSP EWSR1 dual color break-apart probe (located at 22q12), as shown in Figure S1B. Combing of the karyotype with amplifications of RUNX1T1 and MYC genes identified two 8q amplifications on 22 p (Figure S1C; Table S2). Case 3 demonstrated IGH rearrangement at diagnosis and relapse. Moreover, the GLP IGH probe displayed one fusion, two red and two green signals (1F2O2G) in a diploid clone, and 2F2O2G in a tetraploid clone, further confirmed by GSP IGH–CCND3 (Figure S4C, D). The metaphase FISH indicated that the IGH gene translocated to 6 p (distal CCND3) and there was obviously some distance between the IGH and CCND3 genes, with no fusion gene of the IGH–CCND3 genes (Figure S4D).
Identification of gene change
A complex karyotype is the existence of three or more clonal aberrations, and this was detected in 80% (8/10) of patients by FISH in our study. Through the FISH technique, a TP53 deletion was found in three cases: two in ALL and one in AML (3/10). Homologous deletion of P16 was found in Case 3, accompanied by the absence of TP53 (Figure S4E; Table S2). Moreover, P16 amplification on the 9 p locus was detected in Case 4 (Figure S7B, C).
We observed the apparent phenomenon of BCL6 and MYC genes’ amplification and deletion. BCL6 was deleted in four cases and amplified in three cases. Furthermore, MYC was deleted in two cases and amplified in two cases. Three cases had MYC deletion, as well as amplification (Figure 4). Case 10 had classical double t(15;17), and also 3F (10%) and 5F (5%) signals in the MYC gene were found in this patient. Moreover, IGH and PDGFRB genes were apparently commonly seen abnormalities in our study in near-triploidy/tetraploidy-AL (Figure 4; Table S2).
Follow-up and outcomes
Treatment, response, and outcome data are presented in Table 1. Median OS was 11 months. All patients received chemotherapy. Of the ALL patients, Case 1 was an 8-year-old child with a history of thalassemia for 3 years, with detection of mutation in the α-globulin (WS site) and β-globulin (CD117 site). Then the patient developed near-tetraploidy ALL. In addition, the patient responded poorly to chemotherapy, presenting with apparent myelosuppression, drug-induced hepatitis, and secondary infection. After treatment lasting 3 months, the patient’s disease ameliorated, and later, the patient received allogeneic hematopoietic stem cell transplant (HSCT), but relapsed after 9 months and finally died of infection. Cases 2, 3, and 4 had a shorter survival period (range, 3–6 months). Especially, Case 3 initially achieved remission after Vincristine, l-asparaginase, and Prednisone (VLP) with Cytarbine and Darubicin (IA) treatment, but then relapsed quickly. Therefore, we changed the treatment regimen to IA and Mitoxantrone (Novantrone) and Ara-C (NA), but failed to elicit a response. Case 4 was an 88-year-old patient, so a regimen suitable for elderly women was used, but the patient did not respond.
Of the AML patients, cases 5, 6, 7, and 8 relapsed after chemotherapy. Cases 5 and 8 achieved remission after treatment for 6 months. However, 4–5 months later, both patients relapsed, were refractory to treatment, and died of lung infection and intracranial hemorrhage, respectively. Case 6 was diagnosed with AML with a history of postoperative gastric lymphoma for 11 years. The patient received chemotherapy for about 6 months. The patient entered remission and received Chinese medicine. However, 4 months later, relapse occurred and decitabine with Cytarabine, Aclarubicin, and G-CSF were administered, but the patient died of intracranial hemorrhage. Case 7, diagnosed with AML-M2 with ins(21;8)(q22;q24q22), was still alive until the end of follow-up. Despite achieving remission after treatment, the patient relapsed after 7 months. During this period, the RUNX1–RUNX1T1 gene was persistently positive, so the patient failed to achieve molecular complete remission. Hence, various protocols were used without achieving remission. In addition, Case 6 was primary near-tetraploidy, while Cases 5 and 7 were secondary near-tetraploidy, accompanied by diploid clones,8,11 and Case 9 died of pyemia after 2 months. For Case 10, we administered all-trans retinoic acid (ATRA) treatment. However, the child developed fever and back pain symptoms after 10 days, which was considered to be retinoic acid syndrome (differentiation syndrome). After the child was relieved of his syndrome, we treated him with other chemotherapeutic regimens combined with ATRA. The patient underwent CCR for 34 months and was positive for the S-type isoform of PML–RARA at the first diagnosis, but became negative after four rounds of chemotherapy.
Discussion
Because of its rarity, near-triploidy/tetraploidy has been seldom studied. Herein, we have described 10 near-triploidy/tetraploidy-AL cases with gene alterations and chromosomal abnormalities. Notably, the t(8;21)(q22;q22) is a common chromosomal change in diploid AML accompanied by loss of sex chromosomes, which correlates with a favorable prognosis.24–26 However, the incidence of t(8;21) is rare in near-tetraploid patients t(8;21)(q22;q22).24,25,27 Herein, we have reported three novel cases (Cases 5, 6, and 7) of near-tetraploidy AML with double t(8;21)(q22;q22). In particular, by using FISH and RT-PCR, Case 7 was first proven to have an insertion translocation of ins(21;8) (q22;q24q22) in diploid and tetraploid cells. Only 14 cases with diploid ins(21;8) have been reported (Table S3).28–30 Furthermore, some reports had revealed that the occurrences of translocation breakpoints near t(9;22) often involve segmental duplications; but whether this also happened in Case 7 is unknown because whole genome sequencing was not performed.31,32 Despite our best efforts, Case 7 did not acquire hematological and cytogenetic remission during follow-up. We screened five genes (FLT3-ITD, CK-kit D816V, NPM1 [exon12], DNMT3AR882, and CEBPA) and detected a CK-kit D816V mutation. We supposed that the CK-kit D816V mutation may be associated with poor prognosis.33 Case 8 survived for 12 months with a chromosome 7q deletion and trisomy 22. Case 9 survived for 2 months with MLL amplification, which further demonstrated that MLL is closely correlated with poor prognosis.
Furthermore, APL in tetraploidy harboring double t(15;17)(q22;q21) is extremely rare, and only 15 cases have been reported.34,35 The majority of cases were Asians. In our present study, we first reported double t(15;17)(q22;q21) in a child (Case 10) with giant and bizarre myeloblasts and faggot cells filled with granules. In reviewing the15 reported cases, 13 patients with APL attained complete remission and responded favorably to ATRA.34,35 Thus, we combined other chemotherapeutic regimens with ATRA to treat this patient. During the 34 months of follow-up, the patient achieved CCR and PML–RARA was negative after treatment. Hence, allogeneic HSCT was unnecessary in this case. Interestingly, small ratios of genomic aberrations in the patient were found, thereby suggesting that these genomic changes did not affect his prognosis.
Among the four patients with ALL, Case 1 had thalassemia disease. Although this patient was the only patient to receive HSCT, the patient relapsed and died. From this case, we propose that thalassemia has a potential correlation with disease severity. For Case 3, we found IGH rearrangement and detected a homogeneous deletion of p16 by FISH.36,37 VLP with IA, IA, and NA regimens were used for the treatment of this patient, but they failed to improve prognosis. The survival of cases 2 and 4 was very short, at 6 and 3 months, respectively. Moreover, we observed gene amplification in ALL patients. Thus, we propose that multi-amplification might be a cause of unfavorable prognosis and near-triploidy/tetraploidy karyotype, in particular, is a high-risk factor in ALL.38,39 From these cases, it is clear that further improvement in therapy and study on the origins of ALL are vital.39
We found deletions, amplifications, translocations, and rearrangements in the 10 patients by inter/metaphase FISH, and also detected a small portion of gene alterations (BCL6, MYC, and PDFGRB genes) and large region chromosome amplifications (1q+, 8q+, [idic (3q)], and [idic (11q)]), compared with conventional karyotype analysis and microarray. We used FISH to explore how they contributed to the leukemogenic process and demonstrated various different chromosomal and genomic abnormalities (Figure 4; Table S2). The phenomenon showed that tetraploidy originated from the missegregation of chromosomes and chromosome instability, as previously reported.40,41
Conclusion
In summary, the present study clearly showed that near-triploidy/tetraploidy-AL cells may possess distinctive features, not only in morphology and immunophenotypic heterogeneity but also in karyotype instability and in response to therapeutic agents. Hence, a new treatment regimen is urgently needed.
Acknowledgments
The authors would like to thank colleagues from the Department of Hematopathology and the Clinical Cytogenetics Laboratory for their great inputs. They thank Gillian Campbell, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author contributions
RQY collected and analyzed data, interpreted the results, and wrote the manuscript, MHJ provided critical ideas and critically reviewed the manuscript, JZZ provided critical reagent and critical support, HC and JG also contributed to the SPSS software design and analyzed data. YYY and LYS collected and analyzed data and interpreted the results. ZL designed the study, analyzed data, and wrote the manuscript. QL designed the study, performed experiments, and interpreted the results. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Mkrtchyan H, Garcia Ney DR, de Ventura ES, et al. Molecular cytogenetic studies characterize a near-triploid complex karyotype in a child with acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2010;197(1):71–74. | ||
Li L, Li J, Li G, et al. A tetraploid minimally differentiated acute myeloblastic leukemia with extensive erythrophagocytosis: a case report and literature review. Int J Hematol. 2012;96(6):801–805. | ||
Berbari B, Behrens ME, Hathaway LJ. Tetraploidy in acute myeloid leukemia is associated with poor outcomes regardless of the presence of favorable risk features, and requires an allogeneic stem cell transplant in first remission. Blood. 2017;130. | ||
Mühlbacher V, Zenger M, Schnittger S, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes Cancer. 2014;53(6):524–536. | ||
Moorman AV, Harrison CJ, Buck GA, et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): analysis of cytogenetic data from patients treated on the medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECoG) 2993 trial. Blood. 2007;109(8):3189–3197. | ||
Nachman JB, Heerema NA, Sather H, et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood. 2007;110(4):1112–1115. | ||
Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–1548. | ||
Lemez P, Michalová K, Zemanová Z, et al. Three cases of near-tetraploid acute myeloid leukemias originating in pluripotent myeloid progenitors. Leuk Res. 1998;22(7):581–588. | ||
Yamamoto K, Nagata K, Kida A, Tsurukubo Y, Hamaguchi H. CD7+ near-tetraploid acute myeloblastic leukemia M2 with double t(8;21)(q22;q22) translocations and Aml1/ETO rearrangements detected by fluorescence in situ hybridization analysis. Int J Hematol. 2001;74(3):316–321. | ||
Xue Y, Pan Y, Liu Z, Li J, Guo Y, Xie X. Tetraploid or near-tetraploid clones characterized by two 8;21 translocations and other chromosomal abnormalities in two patients with acute myeloblastic leukemia. Cancer Genet Cytogenet. 1996;92(1):18–23. | ||
Xue Y, He J, Wang Y, et al. Secondary near-pentaploidy and/or near-tetraploidy characterized by the duplication of 8;21 translocation in the M2 subtype of acute myeloid leukemia. Int J Hematol. 2000;71(4):359–365. | ||
Goud TM, Al Salmani KK, Al Harasi SM, Al Musalhi M, Wasifuddin SM, Rajab A. Importance of fish combined with morphology, immunophenotype and cytogenetic analysis of childhood/adult acute lymphoblastic leukemia in Omani patients. Asian Pac J Cancer Prev. 2015;16(16):7343–7350. | ||
de Braekeleer E, Basinko A, Douet-Guilbert N, et al. Cytogenetics in pre-B and B-cell acute lymphoblastic leukemia: a study of 208 patients diagnosed between 1981 and 2008. Cancer Genet Cytogenet. 2010;200(1):8–15. | ||
Shuster JJ, Falletta JM, Pullen DJ, et al. Prognostic factors in childhood T-cell acute lymphoblastic leukemia: a pediatric oncology group study. Blood. 1990;75(1):166–173. | ||
Pui CH, Williams DL, Kalwinsky DK, et al. Cytogenetic features and serum lactic dehydrogenase level predict a poor treatment outcome for children with pre-B-cell leukemia. Blood. 1986;67(6):1688–1692. | ||
Jaso J, Thomas DA, Cunningham K, et al. Prognostic significance of immunophenotypic and karyotypic features of Philadelphia positive B-lymphoblastic leukemia in the era of tyrosine kinase inhibitors. Cancer. 2011;117(17):4009–4017. | ||
Biondi A, Rambaldi A, Pandolfi PP, et al. Molecular monitoring of the myl/retinoic acid receptor-alpha fusion gene in acute promyelocytic leukemia by polymerase chain reaction. Blood. 1992;80(2):492–497. | ||
Maruyama F, Stass SA, Estey EH, et al. Detection of AML1/ETO fusion transcript as a tool for diagnosing t(8;21) positive acute myelogenous leukemia. Leukemia. 1994;8(1):40–45. | ||
Schlieben S, Borkhardt A, Reinisch I, et al. Incidence and clinical outcome of children with Bcr/Abl-positive acute lymphoblastic leukemia (ALL). A prospective RT-PCR study based on 673 patients enrolled in the German pediatric multicenter therapy trials ALL-BFM-90 and CoALL-05-92. Leukemia. 1996;10(6):957–963. | ||
Huang L, Wang SA, Dinardo C, et al. Tetraploidy/near-tetraploidy acute myeloid leukemia. Leuk Res. 2017;53:20–27. | ||
Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. | ||
Pui CH, Carroll AJ, Head D, et al. Near-triploid and near-tetraploid acute lymphoblastic leukemia of childhood. Blood. 1990;76(3):590–596. | ||
Reikvam H, Hatfield KJ, Kittang AO, Hovland R, Bruserud Ø. Acute myeloid leukemia with the t(8;21) translocation: clinical consequences and biological implications. J Biomed Biotechnol. 2011;2011(2):1–23. | ||
Trivedi PJ, Brahmbhatt MM, Patel DM, Shukla SN, Patel PS. Acute myeloid leukemia with a masked type of three-way t(8;11;21) revealed by fluorescence in situ hybridizations using AML1–ETO probe. J Assoc Genet Technol. 2014;40(1):11–15. | ||
Onozawa M, Fukuhara T, Nigo M, et al. Insertion (21;8)(q22;q22q22): a masked t(8;21) in a patient with acute myelocytic leukemia. Cancer Genet Cytogenet. 2003;147(2):134–139. | ||
Tian X, Wang Y, Zhao F, et al. A new classification of interphase nuclei based on spatial organizations of chromosome 8 and 21 for t(8;21) (q22;q22) acute myeloid leukemia by three-dimensional fluorescence in situ hybridization. Leuk Res. 2015;39(12):1414–1420. | ||
Johnson RC, Savage NM, Chiang T, et al. Hidden mastocytosis in acute myeloid leukemia with t(8;21)(q22;q22). Am J Clin Pathol. 2013;140(4):525–535. | ||
Specchia G, Albano F, Anelli L, et al. Insertions generating the 5’RUNX1/3’CBFA2T1 gene in acute myeloid leukemia cases show variable breakpoints. Genes Chromosomes Cancer. 2004;41(1):86–91. | ||
Rücker FG, Bullinger L, Gribov A, et al. Molecular characterization of AML with ins(21;8)(q22;q22q22) reveals similarity to t(8;21) AML. Genes Chromosomes Cancer. 2011;50(1):51–58. | ||
Lee JH, Wan TS, Ha JS. Acute myeloid leukemia with a novel t(8;21) variant: paracentric inversion-associated ins(21;8). Leuk Lymphoma. 2014;55(2):441–443. | ||
Paulsson K, Haferlach C, Fonatsch C, et al. The idic(X)(q13) in myeloid malignancies: breakpoint clustering in segmental duplications and association with TET2 mutations. Hum Mol Genet. 2010;19(8):1507–1514. | ||
Albano F, Anelli L, Zagaria A, et al. Genomic segmental duplications on the basis of the t(9;22) rearrangement in chronic myeloid leukemia. Oncogene. 2010;29(17):2509–2516. | ||
Santamaría CM, Chillón MC, García-Sanz R, et al. Molecular stratification model for prognosis in cytogenetically normal acute myeloid leukemia. Blood. 2009;114(1):148–152. | ||
Dalia SM, Horna P, Zhang L. Tetraploidy acute promyelocytic leukemia with double t(15;17)/PML-RARA, a case report with review of literature. Int J Clin Exp Pathol. 2014;7(8):5363–5368. | ||
Kojima K, Imaoka M, Noguchi T, et al. Hypocellular acute promyelocytic leukemia with a tetraploid clone characterized by two t(15;17). Cancer Genet Cytogenet. 2003;145(2):169–171. | ||
Huh J, Mun YC, Yoo ES, Seong CM, Chung WS. Submicroscopic deletions of immunoglobulin heavy chain gene (IgH) in precursor B lymphoblastic leukemia with IgH rearrangements. Ann Lab Med. 2015;35(1):128–131. | ||
Bednarski JJ, Pandey R, Schulte E, et al. RAG-mediated DNA double-strand breaks activate a cell type-specific checkpoint to inhibit pre-B cell receptor signals. J Exp Med. 2016;213(2):209–223. | ||
Charrin C, Thomas X, Ffrench M, et al. A report from the LALA-94 and LALA-SA groups on hypodiploidy with 30 to 39 chromosomes and near-triploidy: 2 possible expressions of a sole entity conferring poor prognosis in adult acute lymphoblastic leukemia (ALL). Blood. 2004;104(8):2444–2451. | ||
Cytogenetic abnormalities in adult acute lymphoblastic leukemia: correlations with hematologic findings outcome. A collaborative study of the group Français de Cytogénétique Hématologique. Blood. 1996;87(8):3135–3142. | ||
Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17(2):157–162. | ||
Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121(Pt 23):3859–3866. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.