Back to Journals » International Journal of General Medicine » Volume 16
Identification of ADAM23 as a Potential Signature for Psoriasis Using Integrative Machine-Learning and Experimental Verification
Authors Yao P, Jia Y, Kan X, Chen J, Xu J, Xu H, Shao S, Ni B, Tang J
Received 24 September 2023
Accepted for publication 15 December 2023
Published 22 December 2023 Volume 2023:16 Pages 6051—6064
DOI https://doi.org/10.2147/IJGM.S441262
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Woon-Man Kung
Pingping Yao,1 Yuying Jia,1 Xuewei Kan,1 Jiaqi Chen,1 Jinliang Xu,1 Huichao Xu,1 Shuyang Shao,1 Bing Ni,2 Jun Tang1
1Department of Dermatology, the First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, 230000, People’s Republic of China; 2Department of Pathophysiology, Third Military Medical University, Chongqing, 400038, People’s Republic of China
Correspondence: Bing Ni; Jun Tang, Email [email protected]; [email protected]
Background: Psoriasis is a common chronic, recurrent, and inflammatory skin disease. Identifying novel and potential biomarkers is valuable in the treatment and diagnosis of psoriasis. The goal of this study was to identify novel key biomarkers of psoriasis and analyze the potential underlying mechanisms.
Methods: Psoriasis-related datasets were downloaded from the Gene Expression Omnibus database to screen differential genes in the datasets. Functional and pathway enrichment analyses were performed on the differentially expressed genes (DEGs). Candidate biomarkers for psoriasis were identified from the GSE30999 and GSE6710 datasets using four machine learning algorithms, namely, random forest (RF), least absolute shrinkage and selection operator (LASSO) logistic regression, weighted gene co-expression network analysis (WGCNA), and support vector machine recursive feature elimination (SVM-RFE), and were validated using the GSE41662 dataset. Next, we used CIBERSORT and single-cell RNA analysis to explore the relationship between ADAM23 and immune cells. Finally, we validated the expression of the identified biomarkers expressions in human and mouse experiments.
Results: A total of 709 overlapping DEGs were identified, including 426 upregulated and 283 downregulated genes. Enhanced by enrichment analysis, the differentially expressed genes (DEGs) were spatially arranged in relation to immune cell involvement, immune-activating processes, and inflammatory signals. Based on the enrichment analysis, the DEGs were mapped to immune cell involvement, immune-activating processes, and inflammatory signals. Four machine learning strategies and single-cell RNA sequencing analysis showed that ADAM23, a disintegrin and metalloprotease, may be a unique, critical biomarker with high diagnostic accuracy for psoriasis. Based on CIBERSORT analysis, ADAM23 was found to be associated with a variety of immune cells, such as macrophages and mast cells, and it was upregulated in the macrophages of psoriatic lesions in patients and mice.
Conclusion: ADAM23 may be a potential biomarker in the diagnosis of psoriasis and may contribute to the pathogenesis by regulating immunological activity in psoriatic lesions.
Keywords: psoriasis, machine learning, differential gene analysis, CIBERSORT, ADAM23
Introduction
Psoriasis is a chronic skin condition caused by skin cells that multiply faster than normal due to immune system dysregulation.1 Psoriasis is classified by the World Health Organization as one of the most serious non-communicable diseases, affecting 2–3% of the global population. Inverse psoriasis (IP), also known as flexural or intertriginous psoriasis, is a variety of plaque psoriasis that involves the body folds, most often the axillary, anogenital, and inframammary ones.2 Clinical variants of psoriasis include plaque, guttate, erythrodermic, and pustular psoriasis. Most psoriasis variants have three main clinical features: erythema, thickening, and scaling.3 Drugs targeting TNF-α, IL-23, and IL-17 and signaling pathways such as JAK/STAT are effective in the clinical management of plaque psoriasis. However, alternate inflammatory pathways may be valid for distinct psoriatic variants. The hallmark of psoriasis is sustained inflammation that leads to uncontrolled keratinocyte proliferation and dysfunctional differentiation. The histology of the psoriatic plaque shows acanthosis (epidermal hyperplasia), which overlies inflammatory infiltrates composed of dermal dendritic cells, macrophages, T cells, and neutrophils.4 The current biological treatments for psoriasis include mainly sJAK inhibitors, ROCK inhibitors, S1P agonists, and AhR agonists.5 The newest therapeutic opportunities include the inhibitors of Janus kinase/tyrosine kinase 2, phosphodiesterase-4 and IL-36 receptor.6 The glutathione-S transferase regulation is also helpful in treating psoriasis.7 Although researchers have found that biologics can effectively relieve clinical symptoms and reduce psoriasis areas and severity index patient scores,8 patients still experience frequent adverse events and psoriasis recurrence.9 Therefore, additional potential biomarkers of psoriasis need to be discovered to provide more possible targets for the treatment of psoriasis.
Psoriasis is an immune disease, and immunity plays a key role in the pathogenesis of psoriasis. Immunological studies in recent decades have shown that both innate and acquired immunity play a key role in the pathogenesis of psoriasis.10,11 For instance, it is well known that the pathogenic factors of psoriasis involve antimicrobial peptides, dendritic cells, tumor necrosis factor-α (TNF-α), interleukin (IL)-23, Th17, IL-17, IL-22, and signal transducer and activator of transcription-3. Although innate and adaptive immunity jointly promote the occurrence and development of psoriasis, the specific mechanism of immunity on psoriasis has not been thoroughly elucidated. Therefore, it is essential to determine how immune cells are involved in the occurrence and development of psoriasis.
Machine learning algorithms have been widely used to diagnose psoriasis and to screen molecular markers associated with the disease.12–15 Single-cell RNA sequencing data have revealed immune cell characteristics in psoriasis subsets,16,17 and the molecular immunological mechanisms underlying psoriasis are yet to be fully elucidated. Therefore, we explored the potential molecular markers of psoriasis using the CIBERSORT method and machine learning algorithms based on omics data, which were further verified using clinical samples. We found ADAM23 may be a novel predictor of prospective psoriasis targets and may contribute to the pathogenesis by regulating immunological activity in psoriatic lesions.
Materials and Methods
Microarray Dataset Collection and Data Preprocessing
The raw gene expression profiles of patients with psoriasis were retrieved from the Gene Expression Omnibus (GEO) database.18 To search for RNA-sequencing profiles in the GEO database, we used the keyword “psoriasis” [all fields]. The selection criteria were as follows: the term “microarray dataset” refers to the microarray dataset of the gene expression profile psoriasis and healthy samples should be included in the microarray dataset; and none of the patients received medicinal treatment. Finally, we narrowed our analysis down to three datasets: GSE30999,19,20 GSE6710,21 and GSE41662.22 The GSE30999 dataset comprises psoriasis cases with positive samples and healthy controls. The GSE30999 dataset was derived using the GPL570 platform and includes 85 non-lesion and 85 psoriasis lesion samples. The GSE6710 dataset was derived using the GPL96 platform and includes 13 lesion skin biopsy samples and 13 non-lesion skin samples. The GSE41662 dataset was derived using the GPL570 platform and includes 24 lesion skin samples and 24 non-lesion skin samples. The GSE41662 dataset was included as an external validation dataset. Additionally, R software was used to conduct quality control analysis and microarray data preprocessing with the Bioconductor package.23
Identification of DEGs
Differentially expressed genes (DEGs) were screened from the GSE30999 and GSE6710 datasets using the “limma” package in R.24 The filter conditions for the DEGs were set as an adjusted p-value < 0.05 and the absolute value of | log2 fold change | > 1. The R software ggplot2 V3.3.5 package was used to create volcano plots, while Pheatmap V1.0.12 was used to plot heatmaps for the top 100 DEGs from each dataset, and Venn Diagram V1.6.20 was used to create Venn diagrams.
Go and KEGG Functional Enrichment Analysis of DEGs
Functional enrichment analysis was performed using the R software cluster Profiler V3.18.1 package25 and Goplot V1.0.2 package26 (p < 0.05 and q < 0.05 were considered statistically significant) to explore the functions and pathways of overlapping DEGs. For all overlapping DEGs, Gene Ontology (GO; using the terms: biological process [BP], cellular component [CC], and molecular function [MF]) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses and visualization were performed.
Screening and Validation of Critical Gene Markers
Four algorithms, including random forests (RF),27 least absolute shrinkage and selection operator (LASSO) logistic regression,28 weighted gene co-expression network analysis (WGCNA)29 and Support Vector Machine Recursive Feature Elimination (SVM-RFE),30 were performed to screen for the key signatures for psoriasis. The R software random Forest V4.7–1 package and R software glmnet V4.1–4 package were used for implementing the RF algorithm and LASSO algorithm, respectively. SVM-RFE is a sequential backward feature elimination method based on SVM learning. The “E1071” and “msvmRFE” packages30 were used to find the optimal hub genes for SVM modeling by deleting feature vectors. Therefore, we used SVM-RFE analysis to screen for core biomarkers at the intersections of the DEGs.
Finally, WGCNA was performed on overlapping DEGs obtained from the GSE30999 and GSE6710 datasets. Based on the scale-free topology criterion, soft-power parameters ranging from 1–20 were selected using the “pickSoftThreshold” function in the WGCNA package. The extracted values were selected to construct an adjacency matrix, then the most appropriate value was selected and the correlation matrix was converted first to an adjacency matrix, then to a topological overlap matrix (TOM). Next, the average linkage hierarchical clustering method was used to cluster the genes based on a TOM, in which the minimum module size was set to 50. Thereafter, modules with similarities were merged. Pearson’s correlation analysis was used to evaluate the association between integrated and eigengene modules in psoriasis.
By intersecting the three sets using these algorithms, the critical biomarkers were identified, and the area under the receiver operating characteristic (ROC) curve (AUC) was used to evaluate the diagnostic performance of the critical biomarkers in the validated dataset GSE41662.
Evaluation and Correlation Analysis of Infiltration-Related Immune Cells
Using CIBERSORT,31 the proportions of 22 types of immune cells were screened, with a p-value < 0.05 as the cutoff, to obtain the immune cell infiltration matrix. Principal component analysis (PCA) clustering of the immune cell infiltration matrix was performed using the “ggplot2” package. The “corrplot” package was used to map and visualize the correlations between the 22 types of infiltrating immune cells. Correlations between infiltrating immune cells and crucial biomarkers were determined using Spearman (non-parametric) correlation analysis.
Acquisition and Hematoxylin and Eosin Staining of Psoriasis Tissue Samples from Humans and Mice
Skin lesion samples from patients with psoriasis were obtained from lesion tissue biopsies of patients with plaque psoriasis. Psoriatic lesions from patients with psoriasis were examined using immunohistochemistry. The study was approved by the Ethics Committee of the First Affiliated Hospital of the University of Science and Technology of China (Anhui, Hefei), and all participants provided written informed consent before enrollment.
The animal experimental protocol was approved by the Animal Ethics Committee of the University of Science and Technology of China, in compliance with the Guidelines for the Ethical Review of Laboratory Animal Welfare, People’s Republic of China National Standard GB/T 35892–2018. Eight male mice from the Experimental Animal Center of the University of Science and Technology of China were maintained at 22–24 °C with 50% humidity, and all mice had free access to food and water. An area of 2.5 × 3 cm was selected on the back of each mouse and carefully shaved. Ten mice were randomly divided into two groups: control (normal group) and experimental (IMQ group). Once a day, 62.5 mg of IMQ ointment (Med-shine Pharma, Chengdu, China) was applied to the prepared back skin of each experimental mouse, and 62.5 mg of Vaseline® was applied to the prepared back skin of each control mouse. The skin in the back modeling area was dehydrated and fixed with 4% paraformaldehyde, embedded in paraffin, and cut into 5-μm paraffin sections. Sections from each group were stained with hematoxylin (Beyotime, China) and eosin (H&E). The sections were observed under a light microscope. At the end of the study, the mice were euthanized to obtain samples from psoriatic lesions after intraperitoneal injection of sodium pentobarbital (60 mg/kg) under anesthesia.
Immunofluorescence Staining
Human psoriasis tissue sections were obtained from paraffin samples. For immunofluorescence staining, the sections were incubated overnight at 4 °C and stained with a polyclonal antibody against ADAM23 (diluted 1:200, Rabbit, Bs-5853R, Bioss, London, UK) and a Cy3-labeled polyclonal secondary antibody (diluted 1:500, A0516, Beyotime, Shanghai, China). Sections were counterstained with 4’,6-diamidino-2-phenylindole (DAPI, C1002). Finally, images were acquired using a fluorescence microscope (KF-FL-020; Jiang Feng, Taichung City, China) and quantified using Image-Pro™ Plus analysis software V8.0 (Media Cybernetics, Rockville, MD, USA). CD68 antibody (diluted 1:300, AFSA006) and fluorescein isothiocyanate were purchased from Yeasen (Shanghai, China).
Western Blot
Psoriasis skin of mice lesions were harvested and lysed on ice with RIPA buffer. Protein samples were analyzed using 10% or 12% SDS-PAGE and transferred onto a polyvinylidene fluoride membrane. The membranes were blocked with 5% non-fat milk for 1 h at room temperature (25 °C), washed three times in TBST, and co-incubated with the primary antibody overnight at 4 °C. The membranes were washed three times with TBST and incubated with horseradish peroxidase-conjugated secondary antibodies (1:1000; ABclonal, Wuhan, China) for 1 h at room temperature. The bands were detected using a western ECL blotting substrate according to the manufacturer’s protocol. The following primary antibodies were used: anti-ADAM23 polyclonal antibody (diluted 1:1000; Bioss) and anti-GAPDH antibody (1:1000; AC001; ABclonal). Densitometric analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The Western blot data from mice are expressed as means ± the standard deviation. Comparisons were performed using an unpaired t-test. Differences were considered statistically significant at two-tailed p-values < 0.05. GraphPad Prism V8.4.3 software (Prism 8 for macOS; San Diego, CA, USA) was used for the statistical analyses and to generate graphs.
Immunohistochemistry (IHC)
The skin sections embedded in paraffin were subjected to immunohistochemical analysis. The sections were subjected to heat treatment in a pressure cooker using citrate buffer (pH 6.0) at 121°C for 10 minutes. The endogenous peroxidase in the tissue sections was blocked with methanol containing 0.3% H2O2. The sections were then incubated with the anti-ADAM23 antibody (diluted 1:500; Bioss). Nuclei were stained with hematoxylin.
Results
Identification of DEGs Between Patients with Psoriasis and Controls Using GEO Datasets
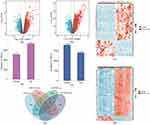
Based on the GSE6710 dataset, a total of 1254 DEGs were identified, of which 518 and 736 genes were downregulated and upregulated, respectively (Figure 1e). A total of 3187 DEGs, of which 1786 and 1401 genes were downregulated and upregulated, respectively, were obtained from the GSE30999 dataset (Figure 1f). The distribution of these DEGs in the GSE6710 and GSE30999 datasets is presented as volcano plots in Figure 1a and b. The top 100 DEGs in the GSE6710 and GSE177477 datasets are shown in heatmaps (Figure 1d and c). A Venn diagram (Figure 1g) was used to show that there were 283 downregulated genes and 426 upregulated genes common in GSE6710 and GSE30999.
GO and KEGG Pathway Analyses of DEGs
GO and KEGG pathway functional enrichment analyses of the 709 overlapping DEGs were conducted, and the results are presented in Figure 2. The results showed that the significantly enriched BPs included the cytokine-mediated signaling pathway, positive regulation of cytokine production, and responses to molecules of bacterial origin (Figure 2a). In the CC category, the DEGs were enriched mainly in the apical part of cell, contractile fiber, and myofibril (Figure 2b). As for the MF category, the most enriched terms were cytokine receptor binding, cytokine activity, and serine-type endopeptidase activity (Figure 2c). Finally, KEGG pathway functional enrichment analysis was conducted on the DEGs. The PPAR signaling pathway, viral protein interactions with cytokines and cytokine receptors, chemokine signaling pathway, IL-17 signaling pathway, and influenza A were regarded as the most significantly enriched pathways (Figure 2d and e).
Screening of Critical Signatures
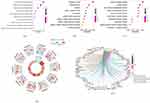
Because differential expressions in GSE30999 were more prominent than those in GSE6710, GSE30999 was used for the subsequent analyses. More specifically, RF, LASSO, WGCNA, and SVM-RFE analyses were performed to screen critical biomarkers, and the results are presented in Figures 3 and 4. The data showed that the mean out-of-band error rate was at a minimum when mtry = 260, which meant that 260 critical genes were selected using the RF algorithms (Figure 3a–c). Moreover, 19 key biomarkers were screened using the LASSO logistic regression algorithm (Figure 3d and e) where the mean-squared error was at a minimum, and 34 key biomarkers were selected with SVM-RFE (Figure 3f). WGCNA was then performed with the default recommended parameters (Figure 4a–d). Twenty notable co-expression modules were identified, of which the turquoise module corresponding to 1388 genes was selected for subsequent investigation because psoriasis was most significantly associated with the turquoise module (maximum p-value = 0.92). Finally, Venn diagrams (Figure 4e) indicated that ADAM23 was the only overlapping gene in these algorithms.
Verification of ADAM23 as a Diagnostic Shared Gene
We validated the critical gene marker ADAM23 by performing ROC analysis on the GSE41662 validated dataset, in which the AUC was 0.932 (Figure 5a), indicating that ADAM23 had a high prediction accuracy for psoriasis. In addition, the expression differences (Figure 5b–d) indicated the notable expression of ADAM23 in lesion and healthy samples. Additionally, ADAM23 was upregulated in psoriasis samples, compared with that in control samples in GSE6710, GSE30999, and GSE41662 (all p < 0.01).
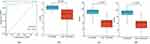 |
Figure 5 Verification of ADAM23 as a diagnostic shared gene. (a) Diagnostic power of ADAM23 in GSE41662. Expression of ADAM23 in the (b) GSE6710, (c) GSE30999, and (d) GSE41662 datasets. |
Immune Infiltration Analysis of Critical Biomarker ADAM23
Considering that the enrichment analysis results showed that the immune response had a significant influence on psoriasis, we performed an immune infiltration analysis of ADAM23 using the CIBERSORT algorithm. PCA was conducted at first, and the results showed that the psoriasis and healthy samples were clearly separated from each other in the plot (Figure 6a). Naïve B cells, follicular helper T cells CD8, T cells, resting CD4+ memory cells, activated NK cells, activated monocytes, and activated dendritic cells expressed positive correlations, and CD4-naïve T cells, neutrophils, and resting mast cells expressed negatively correlations (Figure 6b and c). In addition, as shown in the correlation results (Figure 6d) and box plots (Figure 6e) of the single-cell RNA sequencing data analysis, ADAM23 displayed the strongest positive correlations with monocytes (r = 0.68; p < 0.05) and the most significant negative correlations with resting mast cells (r = −0.54; p < 0.05).
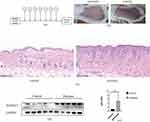
Upregulation of ADAM23 in Mice Psoriasis Samples
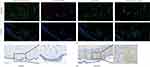
To further validate our findings, we used imiquimod to establish a mouse psoriasis model (Figure 7a and b) and H&E staining to observe a mouse psoriasis model in which psoriatic lesions were significantly thicker than normal skin (Figure 7c). Furthermore, we measured the expression of ADAM23 in mouse psoriatic lesions, and immunoblotting showed that ADAM23 was significantly increased in the skin lesions of psoriatic mice compared to that in the controls (Figure 7d and e). To investigate the relationship between ADAM23 and macrophages, we performed immunofluorescence analysis in human psoriatic lesions and healthy skins. Immunofluorescence staining of psoriasis lesion skins and healthy skins showed colocalization of ADAM23 with CD68-positive macrophages (Figure 8a). Therefore, these results further support the notion that ADAM23, as a potential diagnostic marker of psoriasis, colocalizes with macrophages and potentially contributes to the pathogenesis of psoriatic lesions. Immunohistochemistry showed that ADAM23 was increased in the lesion skins of psoriasis patients compared with healthy controls (Figure 8b).
Discussion
Mounting evidence suggests that the identification of possible biomarkers and targets is critical for the effective treatment of cutaneous inflammatory disorders.32,33 Our study is the first to identify ADAM23 as a potential biomarker for psoriasis. Our results show that ADAM23 may be upregulated in macrophage-rich psoriatic lesions in psoriatic mice as well as in patients with psoriasis. Thus, this is the first report confirming that ADAM23 localizes to macrophages in vivo and that ADAM23 may be a novel potential biomarker for psoriasis. At present, our study demonstrated that ADAM23 is highly expressed in psoriatic lesions compared with healthy skin samples, indicating that ADAM23 may be a diagnosis of psoriasis. In this study, the IHC assay shows that ADAM23 is highly expressed in psoriasis lesion skins, compared with healthy skins, so we suggest that ADAM23 might be a potential biomarker for psoriasis. However, we lack the related evidence to determine whether it could be regarded as a biomarker for early detection or late detection, or the indicator of treatment efficiency. Currently, the diagnosis of psoriasis is mainly based on clinical diagnosis. We propose that high ADAM23 expression could be a potential biomarker for the diagnosis of psoriasis, which support its potential application in clinical practice.
Bioinformatics and machine learning algorithms have expanded our understanding of the mechanisms underlying psoriasis. Many studies have used machine learning algorithms for disease analysis.34,35 In this study, we used four machine learning algorithms, WGCNA, RF, LASSO, and SVM-RFE, to screen key psoriasis genes. The purpose of using multiple machine learning algorithms was to establish a consensus model. The intersection of markers extracted from this algorithm can help to identify more specific psoriasis-related biomarkers from a large amount of data, thereby enhancing the specificity and sensitivity of the biomarker. ADAM23 was selected as a potential signature of psoriasis using four machine learning algorithms. In addition, ROC analysis showed that ADAM23 levels differed significantly between psoriatic lesions and healthy human tissues. ADAM23 maintained a stable performance on other datasets.
Innate immunity and adaptive immunity play key roles in the pathogenesis of psoriasis. An increased number of immune cells (especially dendritic cells and T cells) are present in psoriatic lesions.36 Given the vital role of immunity in psoriasis, we attempted to explore the relationship between psoriasis-related biomarkers and immune cells. We used the CIBERSORT algorithm, and the results showed that ADAM23 was highly expressed on macrophages. ADAM23 was localized to macrophages by single-cell transcriptome analysis of psoriasis data. Therefore, these data provide new insights into the pathogenesis of psoriasis and the immune mechanisms of ADAM23 in the pathogenesis of psoriasis. To confirm that ADAM23 is a key associated biomarker in psoriasis and is correlated with immune cells, immunofluorescence colocalization and Western blot assay were performed on psoriasis lesions in humans. Investigations backed the high expression of ADAM23 in psoriatic lesions, along with the colocalization of ADAM23 with macrophages.
ADAM23 is a member of the brain macrophage-derived chemokine family37 and is a receptor for leucine-rich repeat LGI family member 3 (LGI3). LGI3 increases proinflammatory proteins, including TNF-α, in macrophages.38,39 HaCaT cells, a keratinocyte, can produce LGI3.40 LGI3 is a proinflammatory cytokine secreted by preadipocytes and macrophages.39 Research has shown that psoriasis is regulated by complex interactions between immune cells, such as keratinocytes, dendritic cells, T lymphocytes, neutrophils, macrophages, natural killer cells, mast cells, and other immune cells.41 Therefore, we hypothesized that both macrophages and ADAM23 are involved in the immune pathogenesis of psoriasis. This is consistent with our results from psoriatic lesions in humans and mice.
In addition to its association with macrophages, ADAM23 is involved in the development and differentiation of the nervous system. ADAM23 is a cell surface glycoprotein expressed by neurons in the central nervous system.42 Several members of a disintegrin and metalloproteinase (ADAM) protein family are involved in biological processes ranging from fertilization to myoblast fusion and nerve cell fate determination. ADAM23 plays a pivotal role in various neuronal processes, including modulation of neuronal excitability and differentiation, facilitation of cell adhesion through integrin binding, promotion of neurite outgrowth and differentiation. Additionally, its involvement has been implicated in cancer metastasis and epilepsy.43,44 In psoriasis, peripheral sensory nerves, especially nociceptors, can promote the release of pathogenic cytokines, such as dermal dendritic cell IL-23 and T cell IL-17, through the secretion of calcitonin gene-related peptide neuropeptides.45,46 Recent studies have shown that mesodermal fibroblast subgroups in mouse psoriasis samples promote the abnormal proliferation of nerve axons and neuroimmune interactions.47 Therefore, ADAM23 may be involved in psoriasis by regulating the nervous system, ADAM23 is also associated with neuroinflammation and neurogenesis. Psoriasis is a chronic, psycho-dermatological condition.48 In the skin–brain axis, psoriasis as a chronic stress activates the hypothalamic–pituitary–adrenal (HPA) axis, which can control the immune system via neuroendocrine factors and the autonomous nervous system.49
Therefore, this study might provide new insights into psoriasis and novel ideas for its treatment and diagnosis. To this end, the limitations of this study should be overcome in the future, including the verification of large-scale clinical samples and the mechanisms by which ADAM23 is involved in the pathogenesis of psoriasis.
Conclusions
Our findings revealed, for the first time, that ADAM23 is implicated in the pathogenesis of psoriasis and upregulation of ADAM23 may be a novel biomarker of psoriasis targets. ADAM23 colocalizes with macrophages in psoriatic lesions, suggesting that its effect might be mediated by the immune system.
Abbreviations
DEGs, Differentially expressed genes; TNF-α, tumor necrosis factor-α; IL, interleukin; GEO, Gene Expression Omnibus; GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; RF, random forests; LASSO, least absolute shrinkage and selection operator; WGCNA, weighted gene co-expression network analysis; SVM-RFE, Support Vector Machine Recursive Feature Elimination; TOM, topological overlap matrix; AUC, area under the curve; ROC, receiver operating characteristic; H&E, hematoxylin and eosin; PCA, principal component analysis.
Data Sharing Statement
The datasets generated during and/or analyzed during the current study are publicly available.
Ethics Approval and Informed Consent
The study was conducted in accordance with the declaration of Helsinki. The study was also approved by the Ethics Committee of the First Affiliated Hospital of the University of Science and Technology of China (Anhui, Hefei), number: 2022-N (A)-270 and 2022-N (H)-131, in compliance with the Guidelines for the Ethical Review of Laboratory Animal Welfare, People’s Republic of China National Standard GB/T 35892-2018.
Acknowledgments
We thank the authors of the GSE6710, GSE41662, and GSE30999 datasets for their contribution.
Author Contributions
All authors read and approved the final manuscript. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study received the funding from The National Natural Science Foundation of China (grant no. 82273516) and The Anhui Provincial Key Research and Development Plan of China (grant no. 202004j07020002).
Disclosure
The authors declare that there are no conflicts of interest.
References
1. Walter K. Psoriasis. JAMA. 2022;327(19):1936. doi:10.1001/jama.2022.5270
2. Omland SH, Gniadecki R. Psoriasis inversa: a separate identity or a variant of psoriasis vulgaris? Clin Dermatol. 2015;33(4):456–461. doi:10.1016/j.clindermatol.2015.04.007
3. Armstrong AW, Read C. Pathophysiology, Clinical Presentation, and Treatment of Psoriasis: a Review. JAMA. 2020;323(19):1945–1960. doi:10.1001/jama.2020.4006
4. Rendon A, Schäkel K. Psoriasis Pathogenesis and Treatment. Int J Mol Sci. 2019;20(6):1475. doi:10.3390/ijms20061475
5. Tokuyama M, Mabuchi T. New Treatment Addressing the Pathogenesis of Psoriasis. Int J Mol Sci. 2020;21(20):7488. doi:10.3390/ijms21207488
6. Campione E, Cosio T, Di Prete M, Lanna C, Dattola A, Bianchi L. Experimental Pharmacological Management of Psoriasis. J Exp Pharmacol. 2021;13:725–737. doi:10.2147/JEP.S265632
7. Campione E, Mazzilli S, Di Prete M, et al. The Role of Glutathione-S Transferase in Psoriasis and Associated Comorbidities and the Effect of Dimethyl Fumarate in This Pathway. Front Med Lausanne. 2022;9:760852. doi:10.3389/fmed.2022.760852
8. Al-Hammadi A, Ruszczak Z, Magariños G, Chu CY, El Dershaby Y, Tarcha N. Intermittent use of biologic agents for the treatment of psoriasis in adults. J Eur Academy Dermatol Venereol. 2021;35(2):360–367. doi:10.1111/jdv.16803
9. Gordon KB, Strober B, Lebwohl M, et al. Efficacy and safety of risankizumab in moderate-to-severe plaque psoriasis (UltIMMa-1 and UltIMMa-2): results from two double-blind, randomised, placebo-controlled and ustekinumab-controlled Phase 3 trials. Lancet. 2018;392(10148):650–661. doi:10.1016/S0140-6736(18)31713-6
10. Uppala R, Tsoi LC, Harms PW, et al. “Autoinflammatory psoriasis”-genetics and biology of pustular psoriasis. Cell Mol Immunol. 2021;18(2):307–317. doi:10.1038/s41423-020-0519-3
11. Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32(1):227–255. doi:10.1146/annurev-immunol-032713-120225
12. Lebwohl M, Soliman AM, Yang H, Wang J, Freimark J, Puig L. Impact of PASI response on work productivity and the effect of risankizumab on indirect costs using machine learning in patients with moderate-to-severe psoriasis. J DermatolTreat. 2022;33(4):2094–2101. doi:10.1080/09546634.2021.1919287
13. Gottlieb AB, Mease PJ, Kirkham B, et al. Secukinumab Efficacy in Psoriatic Arthritis: machine Learning and Meta-analysis of Four Phase 3 Trials. J Clin Rheumatol. 2021;27(6):239–247. doi:10.1097/RHU.0000000000001302
14. Meienberger N, Anzengruber F, Amruthalingam L, et al. Observer-independent assessment of psoriasis-affected area using machine learning. J Eur Academy Dermatol Venereol. 2020;34(6):1362–1368. doi:10.1111/jdv.16002
15. Martínez BA, Shrotri S, Kingsmore KM, Bachali P, Grammer AC, Lipsky PE. Machine learning reveals distinct gene signature profiles in lesional and nonlesional regions of inflammatory skin diseases. Sci Adv. 2022;8(17):eabn4776. doi:10.1126/sciadv.abn4776
16. Hughes TK, Wadsworth MH, Gierahn TM, et al. Second-Strand Synthesis-Based Massively Parallel scRNA-Seq Reveals Cellular States and Molecular Features of Human Inflammatory Skin Pathologies. Immunity. 2020;53(4):878–94.e7. doi:10.1016/j.immuni.2020.09.015
17. Liu Y, Wang H, Taylor M, et al. Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Sci Immunol. 2022;7(70):eabl9165. doi:10.1126/sciimmunol.abl9165
18. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012;41(D1):D991–D995. doi:10.1093/nar/gks1193
19. Suárez-Fariñas M, Li K, Fuentes-Duculan J, Hayden K, Brodmerkel C, Krueger JG. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol. 2012;132(11):2552–2564. doi:10.1038/jid.2012.184
20. Correa da Rosa J, Kim J, Tian S, Tomalin LE, Krueger JG, Suárez-Fariñas M. Shrinking the Psoriasis Assessment Gap: early Gene-Expression Profiling Accurately Predicts Response to Long-Term Treatment. J Invest Dermatol. 2017;137(2):305–312. doi:10.1016/j.jid.2016.09.015
21. Reischl J, Schwenke S, Beekman JM, Mrowietz U, Stürzebecher S, Heubach JF. Increased expression of Wnt5a in psoriatic plaques. J Invest Dermatol. 2007;127(1):163–169. doi:10.1038/sj.jid.5700488
22. Bigler J, Rand HA, Kerkof K, Timour M, Russell CB. Cross-study homogeneity of psoriasis gene expression in skin across a large expression range. PLoS One. 2013;8(1):e52242. doi:10.1371/journal.pone.0052242
23. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi:10.1186/gb-2004-5-10-r80
24. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. doi:10.1093/nar/gkv007
25. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
26. Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi:10.1093/bioinformatics/btv300
27. Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. doi:10.1023/A:1010933404324
28. Tibshirani R. Regression Shrinkage and Selection via The Lasso: a Retrospective. J Royal Statistical Society Series B. 2011;73(3):273–282. doi:10.1111/j.1467-9868.2011.00771.x
29. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1):559. doi:10.1186/1471-2105-9-559
30. Yoon S, Kim S. AdaBoost-based multiple SVM-RFE for classification of mammograms in DDSM. BMC Med Inform Decis Mak. 2009;9 Suppl 1(Suppl 1):S1. doi:10.1186/1472-6947-9-S1-S1
31. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
32. Huang J, Feng X, Zeng J, et al. Aberrant HO-1/NQO1-Reactive Oxygen Species-ERK Signaling Pathway Contributes to Aggravation of TPA-Induced Irritant Contact Dermatitis in Nrf2-Deficient Mice. J Immunol. 2022;208(6):1424–1433. doi:10.4049/jimmunol.2100577
33. Ge Y, Luo J, Li D, et al. Deficiency of vitamin D receptor in keratinocytes augments dermal fibrosis and inflammation in a mouse model of HOCl-induced scleroderma. Biochem Biophys Res Commun. 2022;591:1–6. doi:10.1016/j.bbrc.2021.12.085
34. Guo F, Zhu X, Wu Z, Zhu L, Wu J, Zhang F. Clinical applications of machine learning in the survival prediction and classification of sepsis: coagulation and heparin usage matter. J Transl Med. 2022;20(1):265. doi:10.1186/s12967-022-03469-6
35. Yue S, Li S, Huang X, et al. Machine learning for the prediction of acute kidney injury in patients with sepsis. J Transl Med. 2022;20(1):215. doi:10.1186/s12967-022-03364-0
36. Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361(5):496–509. doi:10.1056/NEJMra0804595
37. Elizondo DM, Andargie TE, Marshall KM, Zariwala AM, Lipscomb MW. Dendritic cell expression of ADAM23 governs T cell proliferation and cytokine production through the α(v)β(3) integrin receptor. J Leukoc Biol. 2016;100(5):855–864. doi:10.1189/jlb.2HI1115-525R
38. Kim HA, Park WJ, Jeong HS, et al. Leucine-rich glioma inactivated 3 regulates adipogenesis through ADAM23. Biochim Biophys Acta. 2012;1821(6):914–922. doi:10.1016/j.bbalip.2012.02.010
39. Kim HA, Kwon NS, Baek KJ, Kim DS, Yun HY. Leucine-rich glioma inactivated 3 and tumor necrosis factor-α regulate mutually through NF-κB. Cytokine. 2015;72(2):220–223. doi:10.1016/j.cyto.2014.12.023
40. Kim IW, Jeong HS, Kwon NS, Baek KJ, Yun HY, Kim DS. LGI3 promotes human keratinocyte differentiation via the Akt pathway. Exp Dermatol. 2018;27(11):1224–1229. doi:10.1111/exd.13766
41. Yu J, Zhao Q, Wang X, et al. Pathogenesis, multi-omics research, and clinical treatment of psoriasis. J Autoimmun. 2022;133:102916. doi:10.1016/j.jaut.2022.102916
42. Goldsmith AP, Gossage SJ, ffrench-Constant C. ADAM23 is a cell-surface glycoprotein expressed by central nervous system neurons. J Neurosci Res. 2004;78(5):647–658. doi:10.1002/jnr.20320
43. Hsia HE, Tüshaus J, Brummer T, Zheng Y, Scilabra SD, Lichtenthaler SF. Functions of ‘A disintegrin and metalloproteases (ADAMs)’ in the mammalian nervous system. Cell Mol Life Sci. 2019;76(16):3055–3081. doi:10.1007/s00018-019-03173-7
44. Markus-Koch A, Schmitt O, Seemann S, et al. ADAM23 promotes neuronal differentiation of human neural progenitor cells. Cell Mol Biol Lett. 2017;22(1):16. doi:10.1186/s11658-017-0045-1
45. Riol-Blanco L, Ordovas-Montanes J, Perro M, et al. Nociceptive sensory neurons drive interleukin-23-mediated psoriasiform skin inflammation. Nature. 2014;510(7503):157–161. doi:10.1038/nature13199
46. Cohen JA, Edwards TN, Liu AW, et al. Cutaneous TRPV1(+) Neurons Trigger Protective Innate Type 17 Anticipatory Immunity. Cell. 2019;178(4):919–32.e14. doi:10.1016/j.cell.2019.06.022
47. Cai X, Han M, Lou F, et al. Tenascin C(+) papillary fibroblasts facilitate neuro-immune interaction in a mouse model of psoriasis. Nat Commun. 2023;14(1):2004.
48. Liluashvili S, Kituashvili T. Dermatology Life Quality Index and disease coping strategies in psoriasis patients. Postepy Dermatol Alergol. 2019;36(4):419–424. doi:10.5114/ada.2018.75810
49. Weiglein A, Gaffal E, Albrecht A. Probing the Skin-Brain Axis: new Vistas Using Mouse Models. Int J Mol Sci. 2022;23(13):7484. doi:10.3390/ijms23137484
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.