Back to Journals » Journal of Inflammation Research » Volume 16
Identification of a Novel Target Implicated in Chronic Obstructive Sleep Apnea-Related Atrial Fibrillation by Integrative Analysis of Transcriptome and Proteome
Authors Shen J , Liang J , Rejiepu M, Yuan P, Xiang J, Guo Y , Xiaokereti J, Zhang L , Tang B
Received 5 September 2023
Accepted for publication 21 November 2023
Published 29 November 2023 Volume 2023:16 Pages 5677—5695
DOI https://doi.org/10.2147/JIR.S438701
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tara Strutt
Jun Shen,1,2,* Junqing Liang,1,2,* Manzeremu Rejiepu,1,2,* Ping Yuan,3 Jie Xiang,1,2 Yankai Guo,1,2 Jiasuoer Xiaokereti,1,2 Ling Zhang,1,2 Baopeng Tang1,2
1Xinjiang Key Laboratory of Cardiac Electrophysiology and Cardiac Remodeling, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang, People’s Republic of China; 2Cardiac Pacing and Electrophysiology Department, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang, People’s Republic of China; 3Department of Cardiology, Renmin Hospital, Hubei University of Medicine, Shiyan, Hubei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ling Zhang, Xinjiang Key Laboratory of Cardiac Electrophysiology and Cardiac Remodeling, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang, People’s Republic of China, Tel/Fax +86 18690280833, Email [email protected] Baopeng Tang, Cardiac Pacing and Electrophysiology Department, The First Affiliated Hospital of Xinjiang Medical University, 137 Liyushan Road, Urumqi, Xinjiang, 830000, People’s Republic of China, Tel/Fax +86 13477301883, Email [email protected]
Objective: This study aimed to identify a newly identified target involved in atrial fibrillation (AF) linked to chronic obstructive sleep apnea (COSA) through an integrative analysis of transcriptome and proteome.
Methods: Fifteen beagle canines were randomly assigned to three groups: control (CON), obstructive sleep apnea (OSA), and OSA with superior left ganglionated plexi ablation (OSA+GP). A COSA model was established by intermittently obstructing the endotracheal cannula during exhalation for 12 weeks. Left parasternal thoracotomy through the fourth intercostal space allowed for superior left ganglionated plexi (SLGP) ablation. In vivo open-chest electrophysiological programmed stimulation was performed to assess AF inducibility. Histological, transcriptomic, and proteomic analyses were conducted on atrial samples.
Results: After 12 weeks, the OSA group exhibited increased AF inducibility and longer AF durations compared to the CON group. Integrated transcriptomic and proteomic analyses identified 2422 differentially expressed genes (DEGs) and 1194 differentially expressed proteins (DEPs) between OSA and CON groups, as well as between OSA+GP and OSA groups (1850 DEGs and 1418 DEPs). The analysis revealed that differentially regulated DEGs were primarily enriched in mitochondrial biological processes in the CON-vs.-OSA and OSA-vs.-GP comparisons. Notably, the key regulatory molecule GSTZ1 was activated in OSA and inhibited by GP ablation.
Conclusion: These findings suggest that GSTZ1 may play a pivotal role in mitochondrial damage, triggering AF substrate formation, and increasing susceptibility to AF in the context of COSA.
Keywords: atrial fibrillation, obstructive sleep apnea, transcriptome, proteomic, ganglionated plexi ablation
Introduction
OSA is a prevalent sleep disorder characterized by recurrent episodes of upper airway obstruction during sleep, resulting in disrupted breathing patterns and various physiological changes.1 Extensive evidence has established a link between OSA and AF, an irregular heart rhythm.2 Previous studies have demonstrated the potential benefits of low-level vagal stimulation (LLVS) in reducing AF susceptibility in OSA patients by ameliorating sympathetic hyperactivity and minimizing atrial myocyte injury.3 Additionally, SLGP ablation has shown promise in reducing AF occurrence in COSA patients by suppressing sympathetic overactivity.4 However, the precise underlying mechanisms responsible for the favorable effects of LLVS and SLGP ablation in OSA-related AF remain incompletely understood.
Recent investigations have highlighted the role of OSA in promoting systemic inflammation, reactive oxygen species (ROS) generation, and oxidative stress.5–7 Inflammation and oxidative stress are well-established contributors to AF development.8,9 The pathogenesis of OSA-induced AF likely involves multiple factors, including inflammation, oxidative stress, fibrosis, and autonomic dysfunction.10,11 Despite advances in understanding the mechanisms linking OSA to AF and the utilization of GP ablation, the complex nature of increased AF vulnerability following OSA has not been extensively explored using proteomic and transcriptomic methodologies.12–16 These advanced techniques can provide valuable insights into molecular changes in response to OSA and their role in AF development.
In this study, we evaluated the inducibility of AF in an COSA model. Additionally, we examined the effects of post-ganglionic plexi (GP) ablation through burst pacing and histological examination.3,4,15 To investigate the underlying molecular mechanisms, we employed a comprehensive multi-omics approach, which involved analyzing differentially expressed mRNAs and proteins using atrial proteomic and transcriptomic profiles. Furthermore, we utilized bioinformatics analysis, including gene ontology and pathway analysis, to identify potential therapeutic targets. Our study sheds light on the intricate molecular mechanisms that contribute to the restoration of cardiac autonomic balance following GP ablation in OSA. Moreover, we have identified specific targets and pathways that hold promise for innovative treatments. In summary, our findings provide valuable insights into the molecular mechanisms involved in cardiac autonomic rebalancing and offer potential avenues for the development of novel treatments.
Materials and Methods
Animal Experiment
Approval for all animal studies, including Beagle euthanasia procedures, was obtained from the Animal Research Ethics Committee of Xinjiang Medical University (IACUC201902-K04). The experiments were conducted in strict adherence to the guidelines outlined in the Care and Use of Laboratory Animals of the National Institutes of Health for animal experiments.
Fifteen adult male beagles, weighing an average of 15–18 kg, were sourced from the Experimental Animal Center of South China University of Technology. The dogs were randomly assigned to three groups: OSA (n= 5), which underwent OSA for 12 weeks with sham GP ablation; OSA+GP (n= 5), which also underwent OSA for 12 weeks but received GP ablation at the end of the 8th week through left thoracotomy at the fourth intercostal space; and CTRL (n= 5), which served as a control group receiving sham OSA without GP ablation.
Anesthesia was induced in the adult beagles using a combination of Zoletil (0.1mg/kg; Virbac S.A. France) and xylazine (5 mg/kg; Huamu Animal Health Care Products Co., Ltd., China) via intraperitoneal injection. To maintain anesthesia, a 3% sodium pentobarbital solution was administered as required, with a frequency of every 2 hours. The OSA model was established by obstructing the endotracheal tube at the end of expiration, following the methodology described in our previous experiments.4,5 The duration of intermittent autonomic ventilation was gradually reduced from 9 minutes per week during the initial 4 weeks to 5 minutes per week in the 5th week, and this duration was maintained until the completion of the 12th week.
Consistent with our previous experimental procedures, the pericardium was incised during open-heart surgery at the left fourth intercostal space to expose the SLGP. Subsequent high-frequency stimulation (stimulation frequency: 20 Hz, pulse width: 0.1 ms, stimulation voltage: 0.6–4.5 V) resulted in a 50% increase in the RR interval or a 2:1 atrioventricular block, confirming the accurate positioning of the SLGP. For the ablation of the SLGP, a large-tipped electrode catheter (3.5 mm, Biosense-Webster Inc., Diamond Bar, CA, USA) was used.
AF Inducibility by Programmed Stimulation
The inducibility of AF was consistent with previously described methods. In this study, Beagles underwent open-chest surgery, and a 4-F octopolar catheter (Transonic Systems Inc., New York, USA) was inserted into the right atrium to administer programmed stimuli. To assess the inducibility of atrial arrhythmias, 50 Hz burst pacing was applied for 10 seconds, with 10 pacing intervals of 2 seconds each. AF was defined as the presence of an irregular atrial electrogram lasting more than 1 second, accompanied by an irregular ventricular response. The duration of AF was determined as the average duration of all episodes of AF occurring within a 60-second period per Beagle.
Histology
To examine the histological changes in the left atrium (LA) myocardial tissue, acquired atrial tissue samples were fixed in 4% paraformaldehyde and embedded in paraffin using standard histopathological techniques. The tissues were then sliced into 4 µm sections and subjected to HE staining. Micrographs of the stained sections were captured using a Pixera Pro600EX camera attached to a VANOX-S microscope (Tokyo, Japan). The images were analyzed using ImageJ software (version 1.8.0) to assess the pathological changes observed in the LA.
To investigate the ultrastructural alterations in the LA, transmission electron microscopy was performed using a JEM-1220 microscope (JEOL Ltd., Tokyo, Japan). Briefly, the tissues were fixed with a mixture of 2% glutaraldehyde and 2% paraformaldehyde, stained with 1% uranyl acetate, dehydrated using ethanol, and embedded in epoxy resin.
For GSTZ1 staining, a red fluorescent phalloidin conjugate (ab153995, Abcam) was utilized. Images were captured using a Nikon.80i.JP microscope. A total of 30 images per group, with six repetitions, were taken using the same imaging parameters. The images were analyzed by two pathologists in a double-blind manner using ImageJ (Java) software from the National Institutes of Health, USA.
Atrial Transcriptomic Signatures
The nine Beagle samples provided by Minma Technologies Ltd, an ISO15189 accredited laboratory, underwent RNA extraction, sequencing, and library construction. The dogs were humanely sacrificed using air embolism, and their heart tissues were promptly frozen in liquid nitrogen. Total RNA was extracted using Trizol reagent (Invitrogen, USA) and treated with RNase-free DNase to remove genomic DNA. RNA purity and integrity were assessed through agarose gel electrophoresis and the NanoDrop 2000 spectrophotometer (Thermo). RNA concentration was quantified using the Qubit 2.0, and integrity was assessed using the Agilent 2100.
The Epicenter Ribo-Zero kit (Epicentre, USA) was used to remove ribosomal RNA (rRNA) from the total RNA samples. Fragmentation of enriched RNA was achieved with a fragmentation buffer. The resulting fragments served as templates for synthesizing the initial strand of complementary DNA (cDNA) using random hexamers. The second strand of cDNA was generated using a buffer, dNTPs, DNA polymerase I, and RNase H. The products were purified, amplified via PCR, and combined into a final cDNA library containing both long non-coding RNA (lncRNA) and messenger RNA (mRNA). Paired-end sequencing was performed on the Illumina HiSeq2500 platform.Raw data underwent quality control using Trimmomatic v0.33 and FastQC. Trimmomatic eliminated low-quality RNA-seq reads and splice sequences, while FastQC assessed sequence quality.
Atrial Proteomic Signatures
Proteomic experiments were conducted by PTM Biolabs Inc. (Hangzhou, China) following established protocols. Samples were retrieved from −80 °C storage and pulverized to a fine powder using liquid nitrogen. The powder was then treated with lysis buffer (8 M urea, 1% protease inhibitor, 3 μM TSA, and 50 mM NAM) and subjected to ultrasonication for lysis. After centrifugation, the supernatant was collected for protein concentration assessment using the BCA kit.
To prepare the protein solution for digestion, reduction with dithiothreitol and alkylation with iodoacetamide were performed. Dilution with TEAB reduced the urea concentration. Initial digestion with trypsin was followed by a secondary digestion.Tryptic peptides were dissolved in 0.1% formic acid (solvent A) and loaded onto a custom-made reversed-phase analytical column. A gradient elution system was used, and a constant flow rate was maintained. The peptides were subjected to tandem mass spectrometry using a Q ExactiveTM Plus instrument. Data processing was performed using the Maxquant search engine. Tandem mass spectra were searched against the human UniProt database and the reverse decoy database. Precursor and fragment ion tolerances were set, and identification criteria were applied. In summary, proteomics experiments were conducted, and the resulting data were processed and analyzed following established procedures. The methods employed ensured efficient sample preparation, accurate identification of peptides, and reliable data acquisition.
Bioinformatics Analysis
According to the experimental protocol, the participants were divided into three groups: the control group (CON), the OSA group (OSA), and the OSA+GP group (OSA+GP). Firstly, we conducted an analysis to identify genes and proteins that exhibited differential expression among the groups. Genes and proteins with a fold change >1 and a p-value <0.05 were considered significant differentially expressed genes and proteins (DEPs). The DEGs and DEPs were subjected to annotation against the GO, KEGG, and COG/KOG databases in order to ascertain their respective functions. Subsequently, we quantitatively examined the association between these genes and proteins within the nine-quadrant map. The genes and proteins detected in the transcriptome and proteome, as well as the differential genes and differential proteins, were tallied separately. The overall study process is depicted in Figure 1.
Statistical Analysis
R 4.2.0 software and Adobe Photoshop 2021 were employed in this research. Data were presented as mean ± standard deviation, and comparison between groups was performed using unpaired Student’s t-test.
Results
Increased Susceptibility to AF
Figure 1 depicts the comprehensive workflow of the study, showcasing meticulous experimental protocols and analyses. The study design encompassed three distinct groups: normal, model, and surgical intervention groups. Rigorous analyses were conducted, including sophisticated techniques such as PCA analysis, comparative analysis of differential genes, GO and KEGG enrichment analysis, and cluster analysis. These analytical methods provided a comprehensive exploration of the data, enabling a deep understanding of the underlying patterns and biological mechanisms. Moreover, to ensure the robustness of the findings, rigorous validation experiments were performed, further strengthening the credibility and significance of the study.
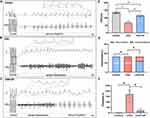
In order to investigate the impact of OSA and GP ablation on the susceptibility to AF, we conducted open-heart surgery on Beagles following our established experimental procedures.3,4 During the surgery, the SLGP was exposed. The results revealed that AF inducibility was significantly elevated in the OSA group under the burst pacing, as well as the AF duration. However, GP ablation exerted an anti-arrhythmic function by reducing AF susceptibility (Figure 2A–C,E,F). As reported in our prior work,3,4 OSA shortened the AERP in the beagles, which were reversed by GP ablation (Figure 2D). It is important to note that further studies with larger sample sizes are necessary to validate these results and determine the statistical significance of the observed differences.
Atrial Enlargements and Atrial Inflammation
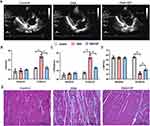
Figure 3A illustrates the typical echocardiographic images of the three groups. Beagles with OSA exhibited significant atrial enlargement, as evidenced by an increased left atrial diameter (LAD). However, in the OSA+GP ablation group, GP ablation partially prevented atrial structural remodeling, as indicated by a significant reduction in atrial enlargement compared to the OSA group (Figure 3B). Furthermore, OSA had a detrimental impact on cardiac function and led to alterations in ventricular structure, including dilation of the left ventricular end-diastolic diameter (LVEDd) and reduced left ventricular ejection fraction (LVEF) (Figure 3C and D).
To investigate the underlying pathological changes associated with atrial remodeling, a histology study was conducted. Examination of the atria of Beagle dogs revealed that OSA resulted in myocardial thickening and significant infiltration of inflammatory cells. Additionally, adipose tissue infiltration was observed in the atrial myocardium. In comparison to OSA group, GP ablation was associated with a less pronounced infiltration of inflammatory cells and adipocytes, suggesting its potential to mitigate the myocardial inflammatory response induced by OSA (Figure 3E).
Taken together, these findings indicate that OSA increases susceptibility to AF and influences the atrial substrate. However, the OSA+GP ablation group exhibits a counteracting effect on the substrate of AF, thereby reducing susceptibility to AF.
Modified Transcriptional Signatures in Beagle Atria
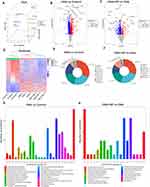
To investigate the underlying mechanisms of increased susceptibility to AF caused by OSA and the reversal of OSA-related AF through GP ablation, transcriptomic and proteomic analyses were conducted on Beagle myocardium. RNA-seq was employed to identify gene expression changes and explore the transcriptome of Beagles with OSA (Supplement 1 and Supplement 2). Both principal component analysis (PCA) and relative standard deviation (RSD) statistical analyses demonstrated the satisfactory reproducibility of gene quantification (Figure 4A and B). The gene expression patterns of these three groups were visualized using heatmaps in Figure 4C. The analysis revealed 2422 differentially expressed genes (DEGs) in the OSA group compared to the control group, with 936 up-regulated and 1486 down-regulated genes (Figure 4D). In comparison to OSA group, the GP group exhibited differential expression of 512 genes, with 411 up-regulated and 101 down-regulated genes (Figure 4E).
GO enrichment analysis was performed to identify significant functional DEGs resulting from OSA and GP ablation treatment. The OSA and control (CON) groups showed enrichment in biological processes such as leukocyte migration, positive regulation of the MAPK cascade, and regulation of inflammatory response. Compared to OSA, the OSA+GP group exhibited enrichment in the regulation of cytokine migration, cytokine chemotaxis, and MAPK cascade processes. Notably, inflammatory factor production, migration, and chemotaxis processes were enriched in all comparisons, indicating the significant impact of GP ablative therapy on the inflammatory response following OSA intervention (Figure 5A–E). KEGG enrichment analysis revealed the activation of the IL-17 signaling pathway, PI3K-Akt signaling pathway, and NF-kappa B signaling pathway by OSA. However, GP ablation disrupted AF susceptibility by modulating these pathways (Figure 6A–E).
Proteomic Signatures Alterations in Beagle Atria
A comprehensive analysis of proteomic profiles was conducted using mass spectrometry, resulting in the identification of 3722 proteins, out of which 3069 were quantifiable (Supplement 3). PCA statistical analyses demonstrated the satisfactory reproducibility of protein quantification (Figure 7A). A comparison between the OSA group and the control group revealed differential expression of 716 down-regulated and 478 up-regulated proteins (Figure 7B). Similarly, a comparison between the OSA+GP group and the OSA group showed differential expression of 383 down-regulated and 128 up-regulated proteins (Figure 7C). The protein expression patterns of these three groups were visualized using heatmaps in Figure 7D.
To gain insights into the subcellular localization of the differentially expressed proteins (DEPs), we performed cellular localization prediction analysis. The results revealed that the primary cellular locations of the DEPs were the cytoplasm, nucleus, extracellular space, and mitochondria (Figure 7E and F). Additionally, we conducted GO taxonomy analysis to assess the functional characteristics of the DEPs in the OSA and OSA+GP groups. The analysis identified a significant involvement of DEPs in metabolic processes, cellular processes, and signaling pathways, including cytoskeletal organization, energy production, and signal transduction mechanisms (Figure 7G and H). Notably, a decrease in the expression of proteins associated with metabolism and signal transduction was observed.
Functional enrichment analysis further highlighted the enrichment of cellular processes, single organ processes, bioregulation, organelles, extracellular regions, cell membranes, binding, and catalytic activity. In the comparison between OSA and CON, we conducted an enrichment analysis of differential proteins. The biological processes (BP) were primarily enriched in areas such as the augmentation of inflammatory factors, calcium ion transport, oxidative stress, and lipid metabolism (Figure 8A). Concurrently, the KEGG pathways were predominantly enriched in PI3K-AKT, MAPK, and calcium ion signaling pathways (Figure 8C). In a similar vein, for the comparison between OSA+GP and OSA, we carried out an enrichment analysis of differential proteins. The BP was primarily enriched in processes such as mitochondrial calcium ion transport, oxidative stress, mitochondrial genesis, and the regulation of autophagy (Figure 8B). The KEGG pathways, on the other hand, were mainly enriched in signaling pathways like NF-KB, oxidative stress, PI3K-AKT and oxidative phosphorylation (Figure 8D). In summary, our analysis revealed distinct patterns of enrichment in both the biological processes and KEGG pathways across the different comparisons. The results revealed prominent enrichment of the IL-17 signaling pathway, PI3K-Akt signaling pathway, and NF-kappa B signaling pathway in all three groups.
Integrated Analysis of Transcriptome and Proteome
To gain a deeper understanding of the biological processes and pathways underlying the reduction of AF susceptibility in dogs with OSA after GP intervention, we performed an integrated analysis of differential genes and differential proteins. Initially, we converted protein IDs to transcript IDs and constructed a correlation matrix based on the transcriptome and proteome data. The correlation plot (Figure 9A–C) displays the top 20 data points representing the correlation between transcriptomics and proteomics. Positive correlations are depicted in orange, while negative correlations are shown in blue, with darker colors indicating stronger correlations. The size of the circles reflects the magnitude of the correlation, with larger circles indicating stronger correlations.
Notably, the up-regulated DEGs in the OSA versus CON group were found to be down-regulated in the OSA+GP group, which is consistent with the proteome-level trends (Figure 9E and H). These findings suggest that OSA may affect multiple common genes, which can be restored through GP ablation. Specifically, 33 DEGs and 55 DEPs were up-regulated in the OSA group compared to the control group but down-regulated in the OSA+GP group (Figure 9D). Additionally, we identified 148 DEGs and 47 DEPs that were down-regulated in the OSA group compared to the CON group but up-regulated in the OSA+GP group (Figure 9G).
The comparative analysis of transcriptional and protein interactions between the OSA and CON groups, as well as the OSA+GP and OSA groups, primarily focused on the regulation of the inflammatory response through the migration and chemotaxis of inflammatory factors. Importantly, the DEGs were predominantly enriched in mitochondrial biological processes in the CON-vs.-OSA and OSA-vs.-GP groups (Figure 9F and I). Furthermore, we confirmed that TSGZ1 and FMO2 were overlapping DEGs/DEPs in both the CON-vs.-OSA and OSA-vs.-GP groups at the transcriptome and proteomic levels (Figure 9J). TSGZ1 expression was up-regulated in the CON-vs.-OSA group but down-regulated in the OSA-vs.-GP group, while FMO2 expression was down-regulated in both the CON-vs.-OSA and OSA-vs.-GP groups (Figure 9K–L). These findings indicate that the key regulatory molecule GSTZ1 is activated during inflammation and inhibited by GP ablation. Overall, these analyses highlight the significance of GSTZ1 as a critical gene involved in mitochondrial biological processes.
Verification of DEGs by TEM and IF Analysis
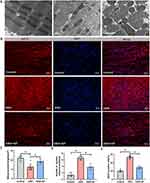
To validate the differential expression of TSGZ1 and examine mitochondrial dysfunction, immunofluorescence staining and transmission electron microscopy (TEM) were performed on canine atrial myocardial tissues. Quantification of mitochondrial lengths and swollen mitochondrial numbers demonstrated that OSA induced the formation of mega-mitochondria, shortened mitochondrial lengths, and increased mitochondrial swelling compared to the control or OSA+GP groups. However, GP ablation effectively mitigated the extent of ultrastructural impairment in myocardial fibers. Furthermore, the proportion of abnormal mitochondria was significantly reduced following GP ablation (Figure 10A and C, D). These findings provide support for the beneficial impact of GP ablation on the preservation of mitochondrial integrity in OSA-related AF.
In addition, the expression of TSGZ1 in atrial myocardial tissues affected OSA was found to be elevated compared to the control group, while its expression was reduced after GP ablation (Figure 10B and E). These results suggest that OSA triggers atrial mitochondrial dysfunction in canines, whereas GP ablation suppresses this activation. Moreover, the association between GSTZ1 and mitochondrial damage highlights its potential role in drug-induced organ toxicity. Further research is needed to unravel the precise mechanisms by which GSTZ1 influences drug metabolism, individual susceptibility to toxic reactions, and mitochondrial function.
Discussion
AF is presently recognized as a prevalent clinical arrhythmia,17 observed in both organic and non-organic heart diseases, posing a significant risk to human well-being.18 Statistical data reveals that the overall population prevalence of AF varies between 0.4% and 1.0%, with incidence rates escalating by a factor of one every decade after reaching 60 years of age.19 Moreover, the prevalence can reach as high as 8% to 10% after the age of 80 years.20 OSA is recognized as an autonomous risk factor for AF.21 Available evidence indicates that the prevalence of AF can range from 40% to 50% in individuals with concurrent OSA, and OSA can elevate the risk of AF by 2 to 3 times.22,23 OSA induces hypoxia, hypercapnia, modified intrathoracic pressure, and autonomic dysfunction within the human body, resulting in structural and functional remodeling of the atria.24,25 In our previous study, we observed an increased susceptibility to AF in the OSA model. Nevertheless, this susceptibility was subsequently diminished after post-GP ablation using burst pacing.3,4,15
The ganglionated plexus plays a crucial role in the rapid electrical excitation of the pulmonary veins, which are associated with AF.26 Cardiac autonomic nerves, rich in adrenergic and cholinergic fibers, are distributed approximately 5 mm from the pulmonary vein-left atrium (PV-LA) junction.27 Prior to innervating the atria, these nerves often converge on specific locations on the atrial surface to form ganglia plexus.28 The GP consists of numerous ganglia and intertwined nerve fiber bundles. In particular, ganglia clusters are commonly found near the openings of the pulmonary vein in the posterior wall of the left atrium, the superior vena cava, and the coronary sinus.29 Therefore, ablation of the circumflex pulmonary vein can disrupt the autonomic nerve connection between the pulmonary vein and the left atrium. Extensive basic and clinical studies conducted by researchers worldwide have highlighted the significant role and mechanism of autonomic nerves in the initiation and maintenance of AF emphasizing the importance of considering their contribution.26–30 Therefore, further investigation is required to elucidate the precise mechanisms and substrates involved in this process.
High-throughput molecular biology techniques, such as transcriptomics and proteomics, have been used to explore complex biological processes.31,32 More and more studies have shown that combining transcriptomics and proteomics techniques to analyse the complex molecular mechanisms underlying the pathogenesis of AF is a promising approach.33–35 In this study, genes and proteins were firstly screened for direct differential expression by OSA stimulation and GP ablation intervention. The study revealed a notable enhancement in the expression of genes and proteins in the OSA group following GP ablation treatment. The integration of transcriptomics and proteomics analysis of the diff-regulated DEGs were mainly enriched in the mitochondrial inner membrane, mitochondrial matrix, mitochondrial respiratory chain complex, and oxidative phosphorylation. Proteomics BP showed that the differentially expressed proteins were mainly enriched in calcium ion transport, oxidative stress, and mitochondrial calcium homeostasis in the CON-vs.-OSA group and OSA-vs.-GP group.
Under pathological conditions, an increased mitochondrial Ca2+ concentration can promote excessive mitochondrial division and affect its function.36 Moreover, it can stimulate the tricarboxylic acid cycle and oxidative phosphorylation, leading to an increase in electron leakage from the respiratory chain and the generation of excessive ROS. It can also induce inappropriate opening of the mPTP, which in turn can induce apoptosis.37 Mitochondria are the core of the body’s energy metabolism and play important roles in oxidative stress, regulation of intracellular calcium homeostasis, intracellular signaling, and the susceptibility of mtDNA to damage in the mechanism of the development and maintenance of AF.38 At the same time, AF is accompanied by abnormal mitochondrial biosynthesis, impaired energy metabolism, homeostasis imbalance of multiple ion channels in cardiomyocytes and mitochondria, and changes in mitochondrial morphology and structure.39–41 Our TEM data showed a marked alteration in the morphological appearance of mitochondria after OSA exposure, which is indicative of mitochondrial damage. Then we quantified mitochondrial lengths and swollen mitochondrial numbers. The results showed that OSA induced the formation of shortened mitochondrial lengths, increased mitochondrial swelling, compared with that of the control or OSA+GP groups. These results suggest that mitochondrial dysfunction may play an important role in the development of AF and mitochondrial damage may accelerate the development of AF.
GSTZ1 is a gene responsible for encoding the enzyme Tyrosine/Tryptophan Ketone Oxygenase, which plays a crucial role in phenylalanine metabolism.42–44 Extensive research has primarily focused on the association between GSTZ1 and diseases, as well as its involvement in drug metabolism and toxic reactions.42–44 In drug metabolism, GSTZ1 plays a significant role in the detoxification and metabolism of various drugs and toxins, including catechol compounds.45–47 Mutations or deficiencies in GSTZ1 can lead to reduced metabolic capacity for these substances, thereby influencing individual responses to medications and increasing the risk of toxic reactions.48–50 Additionally, variations in the expression levels of GSTZ1 have been observed among different tissues and individuals, potentially contributing to differences in drug metabolism and toxic responses.51–54 Therefore, investigating GSTZ1 provides valuable insights into understanding individual variations in drug responses and toxic reactions, forming the foundation for personalized drug therapy. Importantly, no studies to date have reported on the correlation between GSTZ1 and OSA, representing a significant knowledge gap regarding the role of GSTZ1 in OSA.42–44
Furthermore, our study aimed to investigate the correlation between GSTZ1 and OSA, a prevalent sleep disorder associated with various cardiovascular complications, including AF. To achieve this, we employed an OSA model and examined the expression levels of GSTZ1 before and after GP ablation, a common treatment for OSA. Our results revealed a novel finding, demonstrating that GSTZ1 expression was significantly elevated in the OSA model. Interestingly, following GP ablation, the expression of GSTZ1 was downregulated, suggesting a potential role for GSTZ1 in the pathogenesis of OSA-related AF. This finding fills a crucial gap in the understanding of tyrosine metabolism in the context of OSA-associated AF.
To shed light on the possible mechanisms underlying the association between GSTZ1 and AF, we propose that GSTZ1 may contribute to mitochondrial damage, triggering AF matrix formation and increasing susceptibility to AF. Mitochondrial dysfunction has been implicated in the pathogenesis of AF, and GSTZ1, through its involvement in tyrosine metabolism, may play a key role in this process. Further investigations are warranted to elucidate the precise mechanisms by which GSTZ1 influences mitochondrial function and AF development. Taken together, our study provides the first evidence of the correlation between GSTZ1 and OSA. The observed upregulation of GSTZ1 in the OSA model, followed by downregulation post-GP ablation, suggests a potential role for GSTZ1 in OSA-related AF. This finding not only contributes to our understanding of tyrosine metabolism in the context of OSA but also highlights GSTZ1 as a key molecule involved in mitochondrial damage, AF substrate formation, and increased susceptibility to AF.
Limitations
This study has several limitations that should be considered. Firstly, the study was conducted on beagle canines, which may not fully represent the human population. Therefore, further studies on human subjects are needed to validate the findings and establish their relevance to humans. Secondly, the sample size of the study was relatively small, with only fifteen canines divided into three groups. A larger sample size would provide more robust and reliable results. Thirdly, the study focused solely on the correlation between GSTZ1 and OSA-related AF, without investigating other potential factors or mechanisms involved in the development of AF. Future studies should explore these additional factors to gain a more comprehensive understanding of the disease. Additionally, we would like to emphasize that our current research focus is on mitochondria and GSTZ1. We have conducted preliminary validation using immunofluorescence and TEM techniques, but have not yet validated its role at both protein and mRNA levels. In our subsequent studies, we plan to comprehensively validate our findings at the protein, RNA, and cellular levels. Furthermore, we will explore the mechanisms through which GSTZ1 induces mitochondrial dysfunction, leading to increased susceptibility to AF in OSA. Investigating these mechanisms would provide a deeper understanding of the disease pathogenesis. In conclusion, while this study provides valuable insights into the correlation between GSTZ1 and OSA-related AF, further research is needed to address these limitations and fully understand the mechanisms underlying this association.
Conclusion
OSA is a recognized risk factor for AF, causing structural and functional changes in the atria. Our study identified GSTZ1 as a potential gene involved in OSA-related AF, with its expression upregulated in the OSA model and downregulated after GP ablation. GSTZ1 may contribute to mitochondrial damage, increasing susceptibility to AF. This novel finding sheds light on the mechanisms underlying OSA-associated AF and highlights the role of GSTZ1 in mitochondrial dysfunction. Further research is needed to fully understand this association.
Abbreviations
AF, atrial fibrillation; COSA, chronic obstructive sleep apnea; GP, ganglionated plexi ablation; DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSTZ1, Glutathione S-Transferase Zeta 1; RNA-seq, RNA-sequencing; PCA, Principal component analysis.
Ethics Approval and Consent to Participate
Approval for all animal studies, including Beagle euthanasia procedures, was obtained from the Animal Research Ethics Committee of Xinjiang Medical University (IACUC201902-K04). The experiments were conducted in strict adherence to the guidelines outlined in the Care and Use of Laboratory Animals of the National Institutes of Health for animal experiments.
Acknowledgments
We are grateful to the Beagle dogs that made the ultimate sacrifice in our experimental research. We deeply appreciate their contribution and dedication to advancing scientific knowledge. We recognize the ethical considerations associated with animal research and the invaluable insights gained from their involvement. We acknowledge their sacrifice and reiterate our commitment to using alternative methods whenever feasible to reduce animal testing. Additionally, we extend our gratitude to the diligent team members who worked to ensure the accuracy and clarity of this manuscript’s language and translation, significantly enhancing the quality of this work.
Supplementary Materials
Additional tables and figures supporting our findings are available in the Supplementary Materials. For any further inquiries, please contact the corresponding author.
Funding
This work was supported by the National Natural Science Foundation [grant number 82370326, 82260065, 82200360, 81873488], the Natural Science Foundation of Xinjiang Uygur Autonomous Region [grant number 2021D01D16, 2022TSYCCX0098], the Talents Project of Hubei Provincial Department of Education [grant number Q20212102] and Shiyan Municipal Science and Technology Bureau [grant number 21Y49].
Disclosure
All authors declare no competing interests in this work.
References
1. Chan MTV, Wang CY, Seet E, et al. Association of unrecognized obstructive sleep apnea with postoperative cardiovascular events in patients undergoing major noncardiac surgery. JAMA. 2019;321(18):1788–1798. doi:10.1001/jama.2019.4783
2. Tavares L, Rodríguez-Mañero M, Kreidieh B, et al. Cardiac afferent denervation abolishes ganglionated plexi and sympathetic responses to apnea: implications for atrial fibrillation. Circ Arrhythm Electrophysiol. 2019;12(6):e006942. doi:10.1161/CIRCEP.118.006942
3. Guo Y, Xiaokereti J, Meng Q, et al. Low-level vagus nerve stimulation reverses obstructive sleep apnea-Related atrial fibrillation by ameliorating sympathetic hyperactivity and atrial myocyte injury. Front Physiol. 2021;11:620655. doi:10.3389/fphys.2020.620655
4. Zhang L, Guo Y, Xiaokereti J, et al. Ganglionated plexi ablation suppresses chronic obstructive sleep apnea-related atrial fibrillation by inhibiting cardiac autonomic hyperactivation. Front Physiol. 2021;12:640295. doi:10.3389/fphys.2021.640295
5. Lebkuchen A, Freitas LS, Cardozo KHM, et al. Advances and challenges in pursuing biomarkers for obstructive sleep apnea: implications for the cardiovascular risk. Trends Cardiovasc Med. 2021;31(4):242–249. doi:10.1016/j.tcm.2020.04.003
6. Jordan AS, McSharry DG, Malhotra A. Adult obstructive sleep apnoea. Lancet. 2014;383(9918):736–747. doi:10.1016/S0140-6736(13)60734-5
7. Malhotra A, White DP. Obstructive sleep apnoea. Lancet. 2002;360(9328):237–245. doi:10.1016/S0140-6736(02)09464-3
8. Yeghiazarians Y, Jneid H, Tietjens JR, et al. Obstructive sleep apnea and cardiovascular disease: a scientific statement from the American heart association. Circulation. 2021;144(3):e56–e67. doi:10.1161/CIR.0000000000000988
9. Chen W, Cai X, Yan H, Pan Y. Causal effect of obstructive sleep apnea on atrial fibrillation: a Mendelian randomization study. J Am Heart Assoc. 2021;10(23):e022560. doi:10.1161/JAHA.121.022560
10. Huang B, Liu H, Scherlag BJ, et al. Atrial fibrillation in obstructive sleep apnea: neural mechanisms and emerging therapies. Trends Cardiovasc Med. 2021;31(2):127–132. doi:10.1016/j.tcm.2020.01.006
11. Linz D, Linz B, Hohl M, Böhm M. Atrial arrhythmogenesis in obstructive sleep apnea: therapeutic implications. Sleep Med Rev. 2016;26:87–94. doi:10.1016/j.smrv.2015.03.003
12. Fein AS, Shvilkin A, Shah D, et al. Treatment of obstructive sleep apnea reduces the risk of atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2013;62(4):300–305. doi:10.1016/j.jacc.2013.03.052
13. King A. Arrhythmias: CPAP with ablation reduces AF in OSA. Nat Rev Cardiol. 2013;10(7):364. doi:10.1038/nrcardio.2013.78
14. Wilcox I, Chan KH, Dimitri H. Diagnosis and treatment of obstructive sleep apnea is key to achieving optimal results after catheter ablation of atrial fibrillation. J Am Coll Cardiol. 2014;63(6):607–608. doi:10.1016/j.jacc.2013.08.1650
15. Linz D, Ukena C, Mahfoud F, Neuberger HR, Böhm M. Atrial autonomic innervation: a target for interventional antiarrhythmic therapy? J Am Coll Cardiol. 2014;63(3):215–224. doi:10.1016/j.jacc.2013.09.020
16. Buckley U, Rajendran PS, Shivkumar K. Ganglionated plexus ablation for atrial fibrillation: just because we can, does that mean we should? Heart Rhythm. 2017;14(1):133–134. doi:10.1016/j.hrthm.2016.09.001
17. Baman JR, Passman RS. Atrial fibrillation. JAMA. 2021;325(21):2218. doi:10.1001/jama.2020.23700
18. Michaud GF, Stevenson WG. Atrial fibrillation. N Engl J Med. 2021;384(4):353–361. doi:10.1056/NEJMcp2023658
19. Dorian P, Angaran P. Atrial fibrillation. N Engl J Med. 2021;385(4):382–383.
20. Lip GY, Tse HF, Lane DA. Atrial fibrillation. Lancet. 2012;379:9816):648–661. doi:10.1016/S0140-6736(11)61514-6
21. Linz D, McEvoy RD, Cowie MR, et al. Associations of obstructive sleep apnea with atrial fibrillation and continuous positive airway pressure treatment: a review. JAMA Cardiol. 2018;3(6):532–540. doi:10.1001/jamacardio.2018.0095
22. May AM, Van Wagoner DR, Mehra R. OSA and cardiac arrhythmogenesis: mechanistic insights. Chest. 2017;151(1):225–241. doi:10.1016/j.chest.2016.09.014
23. Rossi VA, Stradling JR, Kohler M. Effects of obstructive sleep apnoea on heart rhythm. Eur Respir J. 2013;41(6):1439–1451. doi:10.1183/09031936.00128412
24. Roche F. Arrhythmias and conduction disturbances in obstructive sleep apnoea: the heart of the problem? Eur Respir J. 2013;41(6):1244–1246. doi:10.1183/09031936.00030813
25. Bradley TD, Floras JS. Obstructive sleep apnoea and its cardiovascular consequences. Lancet. 2009;373:9657):82–93. doi:10.1016/S0140-6736(08)61622-0
26. Chugh A. Ganglionated plexus ablation in patients undergoing pulmonary vein isolation for paroxysmal atrial fibrillation: here we go again. J Am Coll Cardiol. 2013;62(24):2326–2328. doi:10.1016/j.jacc.2013.07.055
27. Hanna P, Buch E, Stavrakis S, et al. Neuroscientific therapies for atrial fibrillation. Cardiovasc Res. 2021;117(7):1732–1745. doi:10.1093/cvr/cvab172
28. Katritsis DG, Pokushalov E, Romanov A, et al. Autonomic denervation added to pulmonary vein isolation for paroxysmal atrial fibrillation: a randomized clinical trial. J Am Coll Cardiol. 2013;62(24):2318–2325. doi:10.1016/j.jacc.2013.06.053
29. Driessen AHG, Berger WR, Krul SPJ, et al. Ganglion plexus ablation in advanced atrial fibrillation: the AFACT Study. J Am Coll Cardiol. 2016;68(11):1155–1165. doi:10.1016/j.jacc.2016.06.036
30. Michaud GF, Kumar S. Surgical ganglionic plexus ablation in atrial fibrillation: is all hope lost for the plexus? J Am Coll Cardiol. 2016;68(11):1166–1168. doi:10.1016/j.jacc.2016.07.731
31. Hanash S. Disease proteomics. Nature. 2003;422(6928):226–232.
32. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4):227–232. doi:10.1038/nrg3185
33. Assum I, Krause J, Scheinhardt MO, et al. Tissue-specific multi-omics analysis of atrial fibrillation. Nat Commun. 2022;13(1):441. doi:10.1038/s41467-022-27953-1
34. Wang B, Lunetta KL, Dupuis J, et al. Integrative omics approach to identifying genes associated with atrial fibrillation. Circ Res. 2020;126(3):350–360. doi:10.1161/CIRCRESAHA.119.315179
35. Kornej J, Hanger VA, Trinquart L, et al. New biomarkers from multiomics approaches: improving risk prediction of atrial fibrillation. Cardiovasc Res. 2021;117(7):1632–1644. doi:10.1093/cvr/cvab073
36. Mason FE, Pronto JRD, Alhussini K, Maack C, Voigt N. Cellular and mitochondrial mechanisms of atrial fibrillation. Basic Res Cardiol. 2020;115(6):72. doi:10.1007/s00395-020-00827-7
37. Pool L, Wijdeveld LFJM, de Groot NMS, et al. The role of mitochondrial dysfunction in atrial fibrillation: translation to druggable target and biomarker discovery. Int J Mol Sci. 2021;22(16):8463. doi:10.3390/ijms22168463
38. Fossier L, Panel M, Butruille L, et al. Enhanced mitochondrial calcium uptake suppresses atrial fibrillation associated with metabolic syndrome. J Am Coll Cardiol. 2022;80(23):2205–2219. doi:10.1016/j.jacc.2022.09.041
39. Voigt N, Maack C, Pronto JRD. Targeting mitochondrial calcium handling to treat atrial fibrillation. J Am Coll Cardiol. 2022;80(23):2220–2223. doi:10.1016/j.jacc.2022.09.043
40. Oropeza-Almazán Y, Blatter LA. Mitochondrial calcium uniporter complex activation protects against calcium alternans in atrial myocytes. Am J Physiol Heart Circ Physiol. 2020;319(4):873–881. doi:10.1152/ajpheart.00375.2020
41. Montaigne D, Marechal X, Lefebvre P, et al. Mitochondrial dysfunction as an arrhythmogenic substrate: a translational proof-of-concept study in patients with metabolic syndrome in whom post-operative atrial fibrillation develops. J Am Coll Cardiol. 2013;62(16):1466–1473. doi:10.1016/j.jacc.2013.03.061
42. Nafissi S, Saadat I, Farrashbandi H, Saadat M. Association between three genetic polymorphisms of glutathione S-transferase Z1 (GSTZ1) and susceptibility to schizophrenia. Psychiatry Res. 2011;187(1–2):314–315. doi:10.1016/j.psychres.2010.11.024
43. James MO, Jahn SC, Zhong G, Smeltz MG, Hu Z, Stacpoole PW. Therapeutic applications of dichloroacetate and the role of glutathione transferase zeta-1. Pharmacol Ther. 2017;170:166–180. doi:10.1016/j.pharmthera.2016.10.018
44. Blackburn AC, Matthaei KI, Lim C, et al. Deficiency of glutathione transferase zeta causes oxidative stress and activation of antioxidant response pathways. Mol Pharmacol. 2006;69(2):650–657. doi:10.1124/mol.105.018911
45. Li W, James MO, McKenzie SC, Calcutt NA, Liu C, Stacpoole PW. Mitochondrion as a novel site of dichloroacetate biotransformation by glutathione transferase zeta 1. J Pharmacol Exp Ther. 2011;336(1):87–94. doi:10.1124/jpet.110.173195
46. Rezaei Z, Saadat I, Saadat M. Association between three genetic polymorphisms of glutathione S-transferase Z1 (GSTZ1) and susceptibility to bipolar disorder. Psychiatry Res. 2012;198(1):166–168. doi:10.1016/j.psychres.2011.09.002
47. Yang P, Zeng Q, Cao WC, et al. Interactions between CYP2E1, GSTZ1 and GSTT1 polymorphisms and exposure to drinking water trihalomethanes and their association with semen quality. Environ Res. 2016;147:445–452. doi:10.1016/j.envres.2016.03.009
48. Lantum HB, Baggs RB, Krenitsky DM, Board PG, Anders MW. Immunohistochemical localization and activity of glutathione transferase zeta (GSTZ1-1) in rat tissues. Drug Metab Dispos. 2002;30(6):616–625. doi:10.1124/dmd.30.6.616
49. Alaidaroos NYA, Alraies A, Waddington RJ, Sloan AJ, Moseley R. Differential SOD2 and GSTZ1 profiles contribute to contrasting dental pulp stem cell susceptibilities to oxidative damage and premature senescence. Stem Cell Res Ther. 2021;12(1):142. doi:10.1186/s13287-021-02209-9
50. Jahn SC, Smeltz MG, Hu Z, et al. Regulation of dichloroacetate biotransformation in rat liver and extrahepatic tissues by GSTZ1 expression and chloride concentration. Biochem Pharmacol. 2018;152:236–243. doi:10.1016/j.bcp.2018.04.001
51. Yang F, Li J, Deng H, et al. GSTZ1-1 deficiency activates NRF2/IGF1R axis in HCC via accumulation of oncometabolite succinylacetone. EMBO J. 2019;38(15):e101964. doi:10.15252/embj.2019101964
52. Board PG, Anders MW. Glutathione transferase zeta: discovery, polymorphic variants, catalysis, inactivation, and properties of Gstz1−/− mice. Drug Metab Rev. 2011;43(2):215–225. doi:10.3109/03602532.2010.549132
53. Li J, Wang Q, Yang Y, et al. GSTZ1 deficiency promotes hepatocellular carcinoma proliferation via activation of the KEAP1/NRF2 pathway. J Exp Clin Cancer Res. 2019;38(1):438. doi:10.1186/s13046-019-1459-6
54. Wang Q, Bin C, Xue Q, et al. GSTZ1 sensitizes hepatocellular carcinoma cells to sorafenib-induced ferroptosis via inhibition of NRF2/GPX4 axis. Cell Death Dis. 2021;12(5):426. doi:10.1038/s41419-021-03718-4
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.