Back to Journals » Infection and Drug Resistance » Volume 15
Identification of a Multidrug Resistant Pseudomonas aeruginosa Isolate Harboring Infrequent Red Fluorescence Plasmid from COPD Patient
Authors Zeng Q , Yang X, Li H, Zhang J, Zhang Y, Zhou H, Zhao K
Received 2 August 2022
Accepted for publication 1 December 2022
Published 13 December 2022 Volume 2022:15 Pages 7301—7305
DOI https://doi.org/10.2147/IDR.S383820
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Qianglin Zeng,1 Xiting Yang,1 Heyue Li,2 Jiajie Zhang,1 Yamei Zhang,1 Hui Zhou,1 Kelei Zhao1
1Affiliated Hospital/Clinical College of Chengdu University, Antibiotics Research and Re-Evaluation Key Laboratory of Sichuan Province, School of Pharmacy, Chengdu University, Chengdu, Sichuan, 610106, People’s Republic of China; 2Key Laboratory of Bio-Resources and Eco-Environment, Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, Sichuan, 610064, People’s Republic of China
Correspondence: Kelei Zhao, Affiliated Hospital/Clinical College of Chengdu University, Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province, School of Pharmacy, Chengdu University, No. 2025, Chengluo Avenue, Chengdu, Sichuan, 610106, People’s Republic of China, Email [email protected]
Abstract: Pseudomonas aeruginosa is a notorious Gram-negative opportunistic pathogen that normally causes acute and chronic infections in a wide range of hosts. In this study, a multi-resistant P. aeruginosa isolate L1a harboring an infrequent plasmid with red fluorescence was obtained from the bronchoalveolar lavage fluid of a patient with chronic obstructive pulmonary disease. The results of susceptibility testing and virulence-related phenotypic identification revealed that P. aeruginosa L1a was resistant to levofloxacin, cefepime, aztreonam, and imipenem and showed significantly stronger capacities for swimming and pyocyanin production than the reference strain PAO1. The genome of P. aeruginosa L1a was assembled into one circular chromosome (6,216,913 bp) and one circular plasmid (9111 bp). P. aeruginosa L1a was found to belong to the multilocus sequence type ST549, and serotype O5, and carried 8 drug resistance genes and 18 multidrug efflux pump-related genes in the chromosomal DNA. The plasmid pL1a harbored a tetracycline resistant gene tetA and a functional red fluorescence protein. This study reports a multidrug resistant P. aeruginosa clinical isolate harboring an infrequent red fluorescence plasmid for the first time.
Keywords: Pseudomonas aeruginosa, drug resistance, phenotypic identification, whole-genome sequencing, fluorescence
Chronic obstructive pulmonary disease (COPD) is one of the major public health problems that brings increasing morbidity and mortality worldwide and has become the third leading cause of death following cancers and cardiovascular diseases.1–3 It is considered that smoking is the main causative factor of COPD, especially in elderly people over 60 years old. In addition to the common clinical manifestations such as dyspnea, cough and excessive phlegm, COPD is frequently accompanied by a series of complications, which greatly worsen the lung functions and impose massive healthcare and economic burdens on patients. Therefore, COPD airways are susceptible to being infected by bacterial pathogens because of their poor immune system.4–7
COPD can be classified into stable COPD and acute exacerbation of COPD (AECOPD) according to the progress of the disease and the clinical manifestation of the patients. Especially, AECOPD is mainly caused by bacterial infection and characterized by recurring attacks (1 to 3 times per year), high mortality (23–36% per year), poor prognosis and hard to be permanently cured, and is the critical incident in resulting the rapid decline of lung function and the deaths of patients. Clinical therapy of AECOPD is mainly dependent on the heavy, combined and continued use of antibiotics. Additionally, the treatment methods are differed in dependence of the presence of Pseudomonas aeruginosa, which is a notorious Gram-negative pathogen closely relevant to the recurrence of AECOPD and the readmissions of the patients.1,7–10
P. aeruginosa has a relatively larger genome (≈ 6.3 Mbp) than other common pathogenic bacteria and contains over 5500 genes, including dozens of two-component regulatory genes and membrane transport (efflux pump) systems.11 The complicated and flexible transcriptional network and multidrug resistance mechanisms contribute P. aeruginosa a growth advantage to colonizing a variety of adverse environmental habitats and host tissues, causing acute and chronic infections in immunocompromised patients, and resisting the clearance of host immune system and antibiotics.12,13 The carbapenem-resistant P. aeruginosa has been recognized as Priority 1 (critical) antibiotic-resistant bacteria in the latest global priority list released by the World Health Organization in 2017.14 According to the latest report of China Antimicrobial Surveillance Network (CHINET) in 2021 (www.chinets.com), P. aeruginosa is the third (13.5%) most prevalent bacterial pathogen following Klebsiella pneumoniae (18.6%) and Acinetobacter baumannii (14.1%) among the 114,033 respiratory tract samples collected throughout the country. The resistance of P. aeruginosa isolates (n = 24,035) to imipenem reaches 23%, and to cefepime reaches 9.4%. Here, we report the complete genome of a multidrug resistant P. aeruginosa isolate L1a in China, in an effort to characterize its genomic and functional features, particularly the presence of an infrequent plasmid containing red fluorescence-encoding gene.
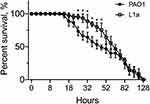
P. aeruginosa L1a was isolated from the bronchoalveolar lavage fluid of a female COPD patient (79-year-old) hospitalized in the Affiliated Hospital of Chengdu University (Sichuan, China). Species identification was performed according to the green pigment of the colony, followed by 16S rDNA sequencing and BLAST against the NCBI database. Susceptibility of P. aeruginosa L1a to the commonly used antibiotics (Sigma-Aldrich, USA) in the clinic was determined by the broth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute.15 The result showed that P. aeruginosa L1a was resistant to levofloxacin (16 μg/mL), cefepime (32 μg/mL), aztreonam (32 μg/mL), and imipenem (8 μg/mL) (Table 1). Compared to the reference strain P. aeruginosa PAO1, P. aeruginosa L1a had significantly stronger swimming motility (about 1.1 folds, p < 0.01) and pyocyanin production (about 2.7 folds, p < 0.01) abilities, but produced moderately lower amount of biofilm (Table 1). Moreover, P. aeruginosa L1a formed a proteolytic ring comparable to P. aeruginosa PAO1 on M9-skim milk plate and could grow on M9-adenosine plate (Table 1), indicating the intact quorum-sensing circuitry of L1a.16 Moreover, the virulence of P. aeruginosa L1a was generally similar to that of PAO1 as determined by the fast-killing assay using Caenorhabditis elegans infection model, albeit L1a killed fewer nematodes than PAO1 between 26 and 45 hours post infection (Figure 1). These results collectively suggested that P. aeruginosa L1a was multidrug resistant and adapted to a planktonic lifestyle in causing acute infections. Correspondingly, P. aeruginosa L1a was isolated as soon as the patient was hospitalized, and the patient passed away in only one month due to severe pulmonary infection.
 |
Table 1 Minimum Inhibitory Concentrations (μg/mL) of Commonly Used Antibiotic Resistance and Virulence-Related Phenotypes of P. aeruginosa L1a |
Genomic DNA of P. aeruginosa L1a was then extracted and subjected to whole-genome sequencing with the Nanopore PromethION platform and Illumina NovaSeq PE150 platform with over 100 × sequence depth. The sequence reads obtained from the two platforms were combined and assembled with Unicycler and SPAdes. Finally, the chromosomal DNA of P. aeruginosa L1a was assembled into one 0gap circular complete sequence with 6,216,913 bp in total length and 66.54% in GC-content. The prediction of genome components including coding sequences (CDSs), interspersed repetitive sequences, non-coding RNAs, Genomics Islands (GIs), Prophage, annotation of CDSs, and the prediction of resistance and virulence genes were performed as described elsewhere.17 The results of genomic analyses revealed that P. aeruginosa L1a is a multilocus sequence type (MLST) ST549 strain according to the 7 loci scheme (identify of 100%). The genome contained 5672 predicted CDSs, 62 tRNA genes, 9 rRNAs, 7 genomic islands, 6 prophages, 198 tandem repeats, 148 minisatellite, and 8 microsatellites. Consistent with the results of susceptibility testing, there were two β-lactamase encoding genes blaOXA-50 and blaPAO, one aminoglycoside resistance gene aph(3’)-IIb, and one tetracycline resistance gene tet(A) in the genome of P. aeruginosa L1a. Additionally, there were also a substantial number of drug resistance determinants encoding resistance to chloramphenicol (catB), fosfomycin (fosA), penicillin (pbp), polymyxin (arnA), and vancomycin (vanB), and multidrug efflux pumps associated with the resistance to aminoglycoside, tigecycline, fluoroquinolone, β-lactam, and tetracycline (mexA-mexB-oprM, emrE), glycylcycline, fluoroquinolone, roxithromycin, and erythromycin (mexC-mexD-oprJ), chloramphenicol and fluoroquinolone (mexE-mexF-oprN), macrolide (macB), and others (mexX-mexW-oprM, mexH-mexI-opmD, and acrA).
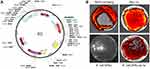
Specifically, P. aeruginosa L1a harbored a circular plasmid (9111 bp in length and 58% in GC-content), designated pL1a, encoding nine proteins, such as TetR family transcriptional regulator, monomeric red fluorescent protein Bc-che1 (mCherry), two hypothetical proteins, replication protein RepA, chromosome-partitioning proteins ParA and ParB, resolvase, and RK2 tetracycline resistance protein TetA (Figure 2A). The result of plasmid classification using PlasmidFinder v1.3 and BLAST showed that pL1a had no significant similarity with any known plasmids or sequences. When the sequence of pL1a was subjected to the software SnapGene, the auto-generated common features of the plasmid showed that pL1a might belong to the pVS1 family according to the backbone reported by Stanisich et al.18 The result of fluorescence test revealed that P. aeruginosa L1a colonies showed red color, indicating the normal expression of pL1a (Figure 2B). When this plasmid was extracted from P. aeruginosa L1a and transformed into fluorescence-negative Escherichia coli DH5α on LB-tetracycline plate (50 μg/mL), the colonies of E. coli DH5α-pL1a successfully produced red fluorescence (Figure 2B). Intriguingly, although the genetic structure of pL1a showed some features of genetic modification for laboratory use, such as the tac promoter and ambiguous multiple cloning site (Figure 2A), there were no documented literatures reporting the application of this plasmid, to the best of our knowledge. Additionally, the patient carrying P. aeruginosa L1a routinely lived in a county of the Tibet Plateau without any laboratories around, and the isolate was identified as soon as she was hospitalized in the Affiliated Hospital of Chengdu University because of AECOPD. Therefore, the origin of the plasmid pL1a is still a mystery.
In conclusion, this is the first report of a multidrug resistant P. aeruginosa clinical isolate harboring an infrequent red fluorescence plasmid. Although it is hard to trace the source of P. aeruginosa L1a and to determine whether the plasmid pL1a is laboratory or wild origin, the present study highlights the susceptibility and complexity of COPD airways in terms of bacterial colonization, and provides a new small and convenient plasmid in labelling Gram-negative bacteria for further research.
GenBank Accession Number
The whole-genome sequence has been deposited in the NCBI database under accession no. PRJNA803822.
Ethical Approval
This study was approved by the Ethics Committee of the Affiliated Hospital of Chengdu University (PJ-2020-021-03). Written informed consent to participate in this study was provided by the participant’s legal guardian/next of kin. This study contains no identifiable features of patients, and complies with the declaration of Helsinki.
Funding
This work was supported by the National Natural Science Foundation of China (31970131), the Open Project of Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province (ARRLKF21-06), the Sichuan Science and Technology Program (2021JDJQ0042), the high-level talent training program of Chengdu University (2081920066), and the innovation foundation of the Affiliated Hospital of Chengdu University (CDFYCX202209).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389:1931–1940. doi:10.1016/S0140-6736(17)31222-9
2. WHO. World Health Organization website; 2021. Available from: http://www.who.int.
3. GOLD. Global initiative for chronic obstructive lung disease. global strategy for diagnosis, management and prevention of chronic pulmonary obstructive disease 2022 report. Available from: https://goldcopd.org/2022-gold-reports-2/.
4. Soriano JB, Kendrick PJ, Paulson KR. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir Med. 2020;8:585–596. doi:10.1016/S2213-2600(20)30105-3
5. Negewo NA, Gibson PG, McDonald VM. COPD and its comorbidities: impact, measurement and mechanisms. Respirology. 2015;20:1160–1171. doi:10.1111/resp.12642
6. Rogliani P, Ritondo BL, Laitano R, et al. Advances in understanding of mechanisms related to increased cardiovascular risk in COPD. Expert Rev Respir Med. 2021;15:59–70. doi:10.1080/17476348.2021.1840982
7. Cavallazzi R, Ramirez J. Community-acquired pneumonia in chronic obstructive pulmonary disease. Curr Opin Infect Dis. 2020;33:173–181. doi:10.1097/QCO.0000000000000639
8. Labaki WW, Rosenberg SR. Chronic obstructive pulmonary disease. Ann Intern Med. 2020;173:ITC17–ITC32. doi:10.7326/AITC202008040
9. Restrepo MI, Babu BL, Reyes LF, et al. Burden and risk factors for Pseudomonas aeruginosa community-acquired pneumonia: a multinational point prevalence study of hospitalised patients. Eur Respir J. 2018;52:1701190. doi:10.1183/13993003.01190-2017
10. Thu PNT, Huong MNT, Thi NT, et al. Combination antibiotic therapy versus monotherapy in the treatment of acute exacerbations of chronic obstructive pulmonary disease: an open-label randomized trial. BMC Infect Dis. 2021;21:1019. doi:10.1186/s12879-021-06687-3
11. Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi:10.1038/35023079
12. Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41:1–20. doi:10.1093/nar/gks1039
13. Zhao K, Li W, Li J, et al. TesG is a type I secretion effector of Pseudomonas aeruginosa that suppresses the host immune response during chronic infection. Nat Microbiol. 2019;4:459–469. doi:10.1038/s41564-018-0322-4
14. World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics; 2017. Available from: http://www.who.int/medicines/publications/global-priority-list-.antibiotic-resistant-bacteria/en/.
15. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 25th informational supplement. In: CLSI Document. Clinical and Laboratory Standards Institute; 2015:M100–S25.
16. Zhao K, Huang T, Lin J, et al. Genetic and functional diversity of Pseudomonas aeruginosa in patients with chronic obstructive pulmonary disease. Front Microbiol. 2020;11:598478. doi:10.3389/fmicb.2020.598478
17. Fang J, Jiao X, Cheng H, et al. Draft genome of a multidrug and multi-heavy metal resistant Vibrio parahaemolyticus ST165 strain of Penaeus vannamei from seawater farms in Zhejiang, China. J Glob Antimicrob Resist. 2021;26:323–325. doi:10.1016/j.jgar.2021.07.015
18. Stanisich VA, Bennett PM, Richmond MH. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J Bacteriol. 1977;129:1227–1233. doi:10.1128/jb.129.3.1227-1233.1977
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.