Back to Journals » Cancer Management and Research » Volume 12
Identification by Comprehensive Bioinformatics Analysis of KIF15 as a Candidate Risk Gene for Triple-Negative Breast Cancer
Authors Sheng J , Li C , Dong M, Jiang K
Received 9 May 2020
Accepted for publication 29 October 2020
Published 1 December 2020 Volume 2020:12 Pages 12337—12348
DOI https://doi.org/10.2147/CMAR.S262017
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Jiayu Sheng, Chunyang Li, Mengting Dong, Ke Jiang
Department of Breast Diseases, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, People’s Republic of China
Correspondence: Ke Jiang
Department of Breast Diseases, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, 110 Ganhe Road, Shanghai 200437, People’s Republic of China
Tel +86 21-65161782
Email [email protected]
Background: Previous studies have shown that kinesin family proteins (KIFs) play an indispensable roles in several types of cancer. However, the expression and clinical significance of KIFs in triple-negative breast cancer remain unclear.
Methods: In this study, the role of KIF15, including gene expression analysis, methylation characteristic, CNV characteristic, and miRNA target regulation, was evaluated using multiple bioinformatic tools based on TCGA database. Quantitative real-time PCR and Western blot were used to determine the expression level of KIF15 in triple-negative breast cancer cell lines. Then, functional experiments were employed to explore the effects of KIF15 on tumor growth and metastasis in triple-negative breast cancer.
Results: Our data showed that KIF15 was significantly upregulated in triple-negative breast cancer (TNBC). Functionally, downregulation of KIF15 significantly facilitated apoptosis and G2/M phase arrest, and inhibited the migration and invasion of TNBC cells. The mechanism of action of KIF15 was closely related to DNA replication checkpoint and cell cycle regulation in TNBC based on GSEA. In addition, bioinformatics analysis demonstrated that high expression of KIF15 in TNBC was correlated with copy number aberration and DNA methylation levels.
Conclusion: Our findings suggest that KIF15 is a novel oncogene in TNBC and provide us a strong evidence that it might be served as a potential clinical target and biomarker in triple-negative breast cancer.
Keywords: kinesin family proteins, bioinformatics analysis, biomarker, triple-negative breast cancer
Introduction
Triple-negative breast cancer (TNBC) refers to breast cancer with negative expression of estrogen receptor (ER) and progesterone receptor (PR) and low expression of human epidermal growth factor receptor (HER2).1,2 Hormone receptor-negative breast cancer is characterized by low differentiation and high histological grade, which is closely related to high recurrence rate, short overall survival, and insensitivity to endocrine therapy.3 However, the prognosis of hormone receptor-negative patients is not very poor; several reports indicated that the prognosis of some patients with medullary cancer is relatively good, suggesting that it is related to the heterogeneity of the tumors.4,5
TNBC is a subtype of breast cancer that lacks clinical therapeutic approaches due to the lack of corresponding therapeutic receptors and poor therapeutic efficacy of endocrine therapy and the molecular targeting of the drugs.6,7 Studies have shown that TNBC is characterized by high proportion of young patients, high histological grade, high rate of axillary lymph node metastasis, low disease-free survival (DFS) and low overall survival (OS) compared with those in non-TNBC.8 TNBC poses a huge challenge for clinical treatment increasing the attention of oncologists to this disease.9 TNBC is the result of interaction of multiple factors and multiple targets. Because of the heterogeneity of TNBC genotype, there are substantial individual differences in its histological morphology, treatment response, and prognosis. The mechanism of TNBC occurrence is still being investigated. At present, many mechanisms of action remain unclear, endocrine and bio-targeted therapies have few options, and main clinic treatments include taxanes and platinum-based chemotherapy with relatively poor efficacy. The molecular mechanism of TNBC remains unclear; however, genetics, reproductive hormones, age and other factors are associated with occurrence and development of TNBC. Activation of oncogenes and inactivation of tumor suppressor genes can induce breast cancer. Numerous studies have shown that a variety of transcription factors play an indispensable role in tumorigenesis and development.10–13 Transcription factors can specifically control the expression of a large number of the downstream genes regulated by the upstream signals to eventually determine the fate of the cells.14,15
Kinesin superfamily proteins (KIFs) are a class of molecular motors that hydrolyze ATP to produce energy to transport intracellular vesicles, organelles, RNA, and other substances.16,17 Kinesin family member 15 (KIF15) is a member of the kinesin-12 subfamily, also known as HKLP2, which is another tetrameric spindle motor in addition to kinesin-5.18 Studies19–21 have shown that KIF is related to the development of breast, lung, and pancreatic cancer. However, changes in the expression of other members of the KIF family in TNBC and the impact of these changes on the prognosis of patients have not been reported in detail. This study is the first to comprehensively analyze the transcriptional level and prognostic significance of KIF15 in 35 human cancers using an online TCGA analysis database. The results of bioinformatics analysis and functional experiments suggest that KIF15 is a novel oncogene in TNBC and that KIF15 may be a clinical biomarker of TNBC.
Methods
Use of the GEPIA Database for Data Extraction
GEPIA (http://gepia.cancer-pku.cn/) contains the RNA sequencing expression data of 9736 tumor and 8587 normal samples of 33 malignant tumors from TCGA and GTEx.22 In this study, the GEPIA database was used to estimate the differences in KIF15 expression in various tumor types by the “Correlation Analysis” module.
LinkedOmics Data Analysis
After logging in to LinkedOmics database homepage,23 “TCGA_BRCA” was selected as cancer type, and “HiSeq RNA” was selected to retrieve the dataset. The input gene was “KIF15”, and “Clinical” was selected for the target dataset. Finally, “Non-parametric t-test” was selected as statistical method; the query was submitted and the results of the analysis were downloaded at P < 0.05. LinkedOmics was used to analyze the correlation between the changes in KIF15 mRNA levels, KIF15 copy number, and methylation.
Gene Set Enrichment Analysis (GSEA)
GSEA software was used (http://www.broadinstitute.org/gsea/index.jsp). The RNA sequence data of the TCGA-TNBC cohort were analyzed and divided into the low or high KIF15 expression groups according to the median expression of KIF15. The purpose of GSEA analysis was to avoid missing genes and pathways that play a key role in the screening of differentially expressed genes. The absolute value of the normalized enrichment score (NES) >1 and the nominal p value <0.05 were considered as the thresholds of statistical significance.
Cell Culture and Transfection
Human TNBC cell lines MDA-MB-231, MDA-MB-468, BT-549, MDA-MB-436, and HCC-1937 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). MDA-MB-231 and stable KIF15-knockdown MDA-MB-231 cells were cultured in DMEM high glucose medium (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum. MDA-MB-436 and MDA-MB-468 cells were cultured in L-15 medium containing 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, USA). BT-549 and HCC-1937 cells were cultured in RPMI 1640 medium (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum. These five cell lines were incubated in a humidified incubator with 5% CO2 at 37°C. The culture medium was replaced every 2 days, and cells in logarithmic growth phase were selected for the experiments. At 90% confluence, the cells were passaged (1:3) or used in the experiments.
The cells were seeded into 6-well plates (2×103 cells/well); after 24 h of culture, the cells were supplemented with pLOV-CMV-shRNA (interference with KIF15 expression) and pLOV-CMV-GFP (negative control) lentiviruses, at a multiplicity of infection (MOI) of 30 in combination with 5.0 μg/mL polybrene (Gibco). After 24 h, the medium was changed, and the cells were passaged in the presence of puromycin (Gibco) to establish stable infected cells. The control shRNA sequence was 5′-TCCACGTAACTCCGAGAAT-3′ and the KIF15-specific shRNA sequence was 5′-TCCAGCATGAATCCCGAAT-3′.
Cell Cycle Assay by Flow Cytometry
Cells in logarithmic growth phase (104) were inoculated into 24-well plates and cultured for specific time for the experiments. The cells were centrifuged at 800 rpm for 5 min, and cell precipitate was collected. Then, the supernatant was discarded and precooled PBS was used to wash the cells twice. Precooled 75% ethanol was added, and the cell precipitate was fixed at 4°C for at least 4 h. Cells were centrifuged at 1500 rpm for 5 min; the supernatant was discarded, and the pellet was washed with 3 mL PBS once; 400 μL PI solution and 100μL RNase A (100μg/mL) were added, and the samples were incubated at 4°C in the dark for 30 min. The cells (20,000 to 30,000) were detected by standard methods, and the results were analyzed by the ModFit software. All experiments were performed three times.
Apoptosis Assay by Flow Cytometry
MDA-MB-231 cells were seeded into 6-well plates (2×105 cells/well) and cultured for 24 h. After digestion with trypsin, the cells were placed in a centrifuge tube and centrifuged at 2000 rpm for 5 min, and the supernatant was removed. Phosphate buffered saline (PBS) was used to wash the cells 3 times. Cells were resuspended in 400 μL binding buffer. Annexin V-FITC (5 μL) was added, and the samples were incubated at 4°C in the dark for 20 min. Apoptosis of the cells was assessed by flow cytometry after the addition of 5μL of PI for 5 min before the assay. The experiments were repeated three times.
Scratch Wound Assay
MDA-MB-231 cells were seeded in 6-well plates (2×105 cells/well) and cultured for 24 h. The back side of the plate was marked with a marker pen and a ruler was used to draw even horizontal lines approximately every 0.5–1 cm across the well. The cells (5×105) were seeded into the wells. On the next day, a straight line was drawn with a pipette tip perpendicular to the line on the bottom. The cells were washed with PBS 3 times to remove the detached cells, and serum-free medium was added. The dishes were placed in a 37°C, incubator with 5% CO2 and imaged at 0, 6, 24, and 48 h. All experiments were repeated three times.
Transwell Assay
MDA-MB-231 cells were seeded into 6-well plates (105 cells/well) and cultured for 24 h. Cells were serum-starved for 12–24 h before preparation of cell suspension to remove the serum. Cell suspension (100μL) was added to a Transwell chamber (Costar, USA). In the lower chamber of a 24-well plate, 600μL of medium containing 20% FBS was added, and the samples were incubated for 12–48 h. The Transwell chamber was removed, and the medium inside it was discarded. The chamber was washed twice with calcium-free PBS, fixed with methanol for 30 min, and dried. After 0.1% crystal violet staining for 20 min, the upper side of the membrane containing unmigrated cells was gently wiped with a cotton swab and washed with PBS 3 times. The cells on the bottom side on the membrane were then observed in five fields of view at 400-fold magnification using a microscope. All experiments were performed three times.
Western Blot
Cells were lysed in protein lysis buffer, vortexed on ice for 10 min (vortexing for 30 s and incubation on ice for 15 s) and centrifuged at 4°C at 12,000 rpm for 5 min. The supernatant (total 20 μg of protein) was mixed with loading buffer and boiled for 5 min. Subsequently, 5% stacking and 10% separation gel was prepared for electrophoresis. After electrophoresis at 120 V for 60 min and 350 mA for 60 min, the proteins were transferred on the PVDF membranes. Then, the membranes were incubated with primary anti-KIF15 (#ab272615, Abcam, 1:1000) and anti-vinculin antibodies (#ab219649, Abcam, 1:500) and with secondary antibodies. Relative grayscale intensity of the bands was analyzed using the ImageJ software. Tubulin was used as the internal standard.
Real-Time PCR (RT-PCR)
Total cellular RNA was extracted using TRIzol reagent (Invitrogen, USA) and stored in a −80°C freezer. The extracted RNA was subjected to reverse transcription using a cDNA first-strand synthesis kit (TaKaRa, PrimeScript RT Master Mix, RR036A). Reversed transcription was performed at 37°C for 15 min and 85°C for 5 s; the cDNA was stored at 4°C. PCR amplification of the target genes and the internal reference GAPDH was carried out using SYBR Premix Ex Taq II, RR820A (Takara, Dalian, China). Primer sequences of KIF15 were as follows: forward, 5ʹ-CTCTCACAGTTGAATGTCCTTG-3ʹ, and reverse: 5ʹ-CTCCTTGTCAGCAGAATGAAG-3ʹ. Primer sequences of GAPDH were as follows: forward, 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ, and reverse: 5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ. RT-PCR was performed at 95°C for 30 s, 95°C for 5 s, and 60°C for 30 s, for a total of 40 cycles. Expression level of the target gene was calculated by the 2–ΔΔCt method. All experiments were performed three times.
Statistical Analysis
The differences in KIF15 expression in cancer and normal tissues were analyzed by
Results
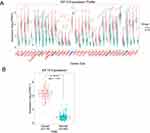
KIF15 is Abnormally Expressed in Various Types of Human Pan-Cancer
In this study, the expression profiles and prognostic value of KIF15 were comprehensively analyzed in 35 cancer types based on the online TCGA analysis database. KIF15 was found to be abnormally expressed in 30 types of cancers, including adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), and colon adenocarcinoma (COAD) (Figure 1A). These data indicate that KIF15 is abnormally expressed in various types of human cancer. Specially, the expression level of KIF15 in the triple-negative breast cancer tissues was higher than that in the normal breast tissues (116 TNBC and 290 normal breast tissues); the corresponding statistical difference is shown in Figure 1B.
Expression of KIF15 in TNBC Cells
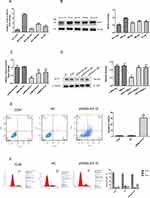
To verify the expression of KIF15 in TNBC, we cultured various TNBC cell lines (HCC-1937, MDA-MB-231, MDA-MB-436, MDA-MB-468, and BT549). The expression of KIF15 in cultured human TNBC cells was detected by real-time quantitative PCR. As shown in Figure 2A, the expression of KIF15 at the mRNA level was significantly higher in MDA-MB-231 cells than that in other cell lines. The protein expression levels of KIF15 in the five cell lines were detected by Western blot. The results were consistent with the expression levels of mRNA as shown in Figure 2B.
Then, MDA-MB-231 cells were selected for infection with lentiviruses in the NC, shKIF15RNA-1 group, shKIF15RNA-2 group, and shKIF15RNA-3 groups. As shown in Figure 2C and D, the effect of knockdown in the shKIF15RNA-1 group was the highest, and a series of subsequent experiments were performed in the shKIF15RNA-1 group.
Effect of KIF-15 on Apoptosis of MDA-MB-231 Cells
To determine the role of KIF15 in apoptosis of TNBC cells, Annexin V/PI double staining was used to evaluate the influence on KIF15 on apoptosis of MDA-MB-231 cells and MDA-MB-231/shKIF15RNA-1 cells. The results showed that apoptosis in MDA-MB-231/KIF-15 knockdown cells in the experimental group was significantly augmented compared with that in the control group, and the difference was statistically significant (P<0.05, Figure 2E). This result suggests that apoptosis and KIF15 expression are significantly correlated, and silencing KIF15 can significantly facilitate apoptosis in MDA-MB-231 cells. Additionally, infected with KIF-15 shRNA induced the G2/M phase arrest in MDA-MB-231 cells (Figure 2F), indicating that cell cycle regulation by KIF15 plays an important role in MDA-MB-231 cells.
KIF15-Related Upstream Mechanisms
miRNAs potentially targeting KIF15 in TNBC were then investigated. Five independent online tools (miRMap, miRDB, TargetScan, miTarBase, and miRanda) were used to predict possible miRNAs targeting KIF15, and 10 candidate miRNAs were identified (Figure 3A, Supplementary Table S1). Hsa-miR-135a-5p, hsa-miR-139-5p, hsa-miR-381-3p, hsa-miR-664a-5p, hsa-miR-2115-5p were correlated with KIF15 expression (Figure 3B–F).
LinkedOmics analysis showed that KIF15 overexpression is positively correlated with its copy number aberration in TNBC (r = 0.358, P < 0.01, Figure 3G), and KIF15 overexpression was negatively correlated with the level of DNA methylation of the KIF15 gene (R2 = 0.239, P < 0.01, Figure 3H).
Effect of KIF15 on the Migration and Invasion Ability of MDA-MB-231 Cells
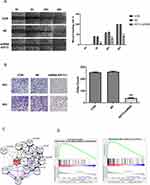
MDA-MB-231 cells infected with lentivirus were used to detect the changes in migration in the three groups at 6 h, 24 h, and 48 h after the scratching. The results showed that the distance between the scratches was significantly lower in the control group; in the KIF15 knockdown group, the distance between the scratches was not significantly lower. There were significant differences between time periods and with the control group (Figure 4A). To explore the influence of KIF-15 on the migration and invasion of MDA-MB-231 cells, the effect of KIF15 knockdown was assessed by the transwell invasion assay. The results indicated that the number of cells invading the lower compartment cells was significantly reduced in MDA-MB-231/KIF15 knockdown compared with that in the control group at 24 h and 48 h (Figure 4B). Thus, cell invasion was significantly reduced after silencing KIF15 in MDA-MB-231 cell.
KIF15-Related Downstream Mechanisms
Investigation of the molecular mechanism of KIF15 in the occurrence and development of TNBC plays an indispensable role in the treatment of the disease and can guide targeted clinical interventions, improving the prognosis of the patients. Genes co-expressed with KIF15 in breast cancer were analyzed using the LinkedOmics database, and a PPI network centered on KIF15 was generated. The proteins associated with KIF15 were NCAPG, KIF23, KIF20A, KIF2C, RACGAP1, CENPE, KIF4A, KIF11, AURKA, and TPX2 (Figure 4C). DNA-binding of transcription factors according to the ChIP-seq data of the ENCODE database predicted the transcription factor relationships regulating KIF15 (Supplementary List S1).
To investigate possible pathways mediated by KIF15, GSEA of the genes coexpressed in TNBC was performed. The results of GSEA showed that KIF15 is mainly involved in DNA damage checkpoint (P=0, FDR=0, NES=2.84) and cell cycle (P=0, FDR=0, NES=1.96) in TNBC (Figure 4D). The top 20 pathways with the most obvious up-regulation and down-regulation are listed in Tables 1 and 2.
 |
Table 1 The GSEA Analysis Results Showed That the First 20 Pathways Were Significantly Enriched |
 |
Table 2 The First 20 Pathways Were Significantly Enriched in GSEA Analysis |
Discussion
Various factors such as genetics, age, and reproductive hormones are significantly associated with the occurrence and development of breast cancer. Overall, activation of the oncogenes and inactivation of the tumor suppressor genes can induce breast cancer. Mutations in the tumor suppressor genes, such as BRCA1, and activation of the oncogenes, such as ERα, Her-2, and beta-catenin, can facilitate breast cell proliferation, survival, and metastasis.24 Transcription factors integrate the upstream signals and specifically control the expression of numerous downstream genes thus determining the fate of the cells.25,26
Recently, a large number of studies27,28 have shown that KIFs are a conserved class of microtubule-dependent molecular motor proteins with ATPase activity, which are associated with neurodegenerative diseases, metabolic diseases, nephropathy, and other diseases. Additionally, KIFs play an essential role in the development of tumors.29 Studies have demonstrated30,31 that during tumor cell division, abnormally expressed KIFs may affect tumor cell proliferation, invasion, and metastasis by influencing chromosome agglutination and spindle formation. For example, KIF1B facilitates glioma cell migration and invasion by inducing membrane-type matrix metalloproteinases.32 In lung cancer cells, downregulation of KIF23 facilitates apoptosis, and KIF23 is considered a potential therapeutic target for lung cancer patients.33 Chen et al34 reported that some KIF family members are associated with TNM stage in HCC patients, and the expression of KIF15 and Ki-67 in tumor tissues is positively correlated. KIF is involved in essential biochemical processes including mitosis, suggesting that KIF15 may affect cell proliferation by influencing mitosis.
Given that KIF15 is upregulated in TNBC, we investigated the functions and possible mechanisms of KIF15 in breast cancer. GSEA indicated that KIF15 may be involved in DNA replication and cell cycle in TNBC. It is known that DNA replication is an essential biological event in cell proliferation. The in vitro data indicate that KIF15 is involved in the proliferation of TNBC cells. Considering that KIF15 may facilitate the development of TNBC by regulating miRNAs, we focused on exploring the miRNA targets of KIF15.
KIF15 is one of 5 candidate genes, for regulation of the expression by DNA methylation of facilitator CpGI35 and KIF15 is associated with severe liver cancer cases and poor prognosis. However, there are no studies on the regulation of KIF15 by miRNAs. Therefore, the upstream mechanism of KIF15 deserves additional study. The correlation between patient age and differences in KIF15 expression indicates that KIF15 expression may be time-specific. Different TNBC cell lines were originally derived from different tumor patients, and heterogeneity may be the reason for the differences in KIF15 expression. Additionally, this study demonstrated that augmented gene copy number and decreased DNA methylation levels facilitate gene expression, which is in agreement with the regulatory mechanism of gene expression, ie, changes in gene copy number can increase the gene dose or promote gene expression. Genomic hypomethylation is a common feature of cancer cells, and increasing DNA methylation levels to attenuate the oncogene expression can be used to treat cancer.
Conclusion
The combination of bioinformatics analysis and experiments with the cultured cells identified upstream and downstream regulation of KIF15. Our results demonstrated that KIF15 is a novel oncogene in TNBC and may be a potential clinical target and biomarker of human malignancy.
Data Sharing Statement
Data and material will be available upon corresponding author approval. All data sets analyzed in this study are included in the manuscript and the additional files.
Ethics Approval and Consent to Participate
Not Applicable
Author Contributions
All authors contributed significantly to concepts and design, data acquisition or data analysis and interpretation; participated in drafting or critical revision of essential academic content; gave final approval to the upcoming version; and agreed to be responsible for all aspects of work. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was funded by the National Natural Science Foundation of China (82004240), the Scientific Research Program of Shanghai Science and Technology Commission (17401935300), the Shanghai Municipal Health and Family Planning Commission (2018LQ020) and the Shanghai Office of Traditional Chinese Medicine Development (ZY2018-2020-RCPY-2009).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yaqin S, Juan J, Wenfei J, Xiaoxiang GJMC. Therapeutic landscape in mutational triple negative breast cancer. Molecular Cancer. 2018;17(1):99.
2. Jia H, Truica CI, Wang B, et al. Immunotherapy for triple-negative breast cancer: existing challenges and exciting prospects. Drug Resistance Updates. 2017;S1368764617300316.
3. Park JH, Jonas SF, Bataillon G, Criscitiello C, Michiels S. Prognostic value of tumor-infiltrating lymphocytes in patients with early-stage triple-negative breast cancers (TNBC) who did not receive adjuvant chemotherapy. Annals of Oncology. 2019;30:12.
4. Mateo AM, Pezzi TA, Sundermeyer M, Kelley CA, Klimberg VS, Pezzi C. Atypical medullary carcinoma of the breast has similar prognostic factors and survival to typical medullary breast carcinoma: 3976 cases from the national cancer data base. Journal of Surgical Oncology. 2016;114:5.
5. Aksoy A, Odabas H, Kaya S, Bozkurt O, Gumus MJSMJ. Hormone receptor status and survival of medullary breast cancer patients. Saudi Medical Journal. 2017;38(2):156–162.
6. Medina MA, Oza G, Sharma A, Arriaga LG, Ramirez J. Triple-negative breast cancer: a review of conventional and advanced therapeutic strategies. International Journal of Environmental Research and Public Health. 2020;17(6):2078.
7. Diana A, Carlino F, Franzese E, Oikonomidou O, Orditura MJC. Early triple negative breast cancer: conventional treatment and emerging therapeutic landscapes. Cancers. 2020;12(4):819.
8. Agarwal G, Nanda G, Lal P, et al. Outcomes of triple-negative breast cancers (TNBC) compared with non-TNBC: does the survival vary for all stages? World Journal of Surgery. 2016;40(6):1–11.
9. Lehmann B, Bauer J, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767.
10. Liang J, Cui Y, Meng Y, et al. Integrated analysis of transcription factors and targets co-expression profiles reveals reduced correlation between transcription factors and target genes in cancer. Functional & Integrative Genomics. 2018.
11. Li Q, Li J, Dai W, Li Y. Aiim. Differential regulation analysis reveals dysfunctional regulatory mechanism involving transcription factors and microRNAs in gastric carcinogenesis. Artificial Intelligence in Medicine. 2017;77:12–22.
12. Makiko BI, Medicine DJWIRS. The mechanistic basis for chromatin regulation by pioneer transcription factors. Wiley Interdisciplinary Reviews Systems Biology & Medicine. 2018.
13. Malysheva V, Mendoza-Parra MA, Saleem MAM, Gronemeyer HJGM. Reconstruction of gene regulatory networks reveals chromatin remodelers and key transcription factors in tumorigenesis. Genome medicine. 2016;8(1):57.
14. Pajtler K, Wei Y, Okonechnikov K, et al. YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat Commun. 2019;10(1):3914.
15. Liu C, Zhang Y, Huang T, Cai Y. Identification of transcription factors that may reprogram lung adenocarcinoma. Artificial intelligence in medicine. 2017;83:52–57.
16. Liu M, Nadar V, Kozielski F, Kozlowska M, Yu W, Baas P. Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. Journal of Neuroscience. 2010;30(44):14896–14906.
17. Eskova A, Knapp B, Matelska D, et al. An RNAi screen identifies KIF15 as a novel regulator of the endocytic trafficking of integrin. J Cell Sci. 2014;127:2433–2447.
18. Florian S, Mayer T. Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles. Cell Cycle. 2011;10(20):3533–3544.
19. Zou J, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Molecular Cancer Research. 2014;12(4):539–549.
20. Bidkhori G, Narimani Z, Hosseini Ashtiani S, Moeini A, Nowzari-Dalini A, Masoudi-Nejad A. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. Plos One. 2013;8(7):e67552.
21. Yokota K, Sasaki H, Okuda K, et al. KIF5B/RET fusion gene in surgically-treated adenocarcinoma of the lung. Oncology Reports. 2012;28(4):1187–1192.
22. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nuclc Acids Research. 2017;45:W98–W102.
23. Wang Y, Wang F, He J, et al. miR-30a-3p Targets MAD2L1 and regulates proliferation of gastric cancer cells. OncoTargets and Therapy. 2019;12:11313–11324.
24. Alexandrova E, Lamberti J, Saggese P, et al. Small Non-Coding RNA Profiling Identifies miR-181a-5p as a mediator of estrogen receptor beta-induced inhibition of cholesterol biosynthesis in triple-negative breast cancer. Cells. 2020;9(4).
25. Pennarossa G, Gandolfi F, Brevini T. Genetics. All roads lead to rome: the many ways to pluripotency. Journal of assisted reproduction and genetics. 2020;37(5):1029–1036.
26. Kresovich JK, Gann PH, Erdal S, et al. Candidate gene DNA methylation associations with breast cancer characteristics and tumor progression. Epigenomics. 2018. doi:10.2217/epi-2017-0119.
27. Yang W, Tanaka Y, Bundo M, Hirokawa N. Antioxidant signaling involving the microtubule motor KIF12 is an intracellular target of nutrition excess in beta cells. Dev Cell. 2014;31(2):202–214.
28. Inomata H, Nakamura Y, Hayakawa A, et al. A scaffold protein JIP-1b enhances amyloid precursor protein phosphorylation by JNK and its association with kinesin light chain 1. Journal of Biological Chemistry. 2003;278(25):22946–22955.
29. Chen J, Li S, Zhou S, et al. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. Journal of Cancer Research and Therapeutics. 2017;13(4):651–659.
30. Sun X, Jin Z, Song X, et al. Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. Bmc Cancer. 2015;15:961.
31. Yu Y, Feng YJC. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer. 2010;116(22):5150–5160.
32. Chen S, Han M, Chen W, et al. KIF1B promotes glioma migration and invasion via cell surface localization of MT1-MMP. Oncology Reports. 2015.
33. Kato T, Wada H, Patel P, et al. Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer. 2016;92:53–61. doi:10.1016/j.lungcan.2015.11.018
34. Vanneste D, Ferreira V, Vernos IJBST. Chromokinesins: localization-dependent functions and regulation during cell division. Biochemical Society Transactions. 2011;39(5):1154.
35. Matsushita J, Suzuki T, Okamura K, Ichihara G, Nohara K. Identification by TCGA database search of five genes that are aberrantly expressed and involved in hepatocellular carcinoma potentially via DNA methylation changes. Environmental Health and Preventive Medicine. 2020;25(1):31.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.