Back to Journals » Journal of Inflammation Research » Volume 16
Identification and Validation of Signature Genes and Potential Therapy Targets of Inflammatory Bowel Disease and Periodontitis
Authors Xiong Z, Fang Y, Lu S, Sun Q, Huang J
Received 5 July 2023
Accepted for publication 12 September 2023
Published 28 September 2023 Volume 2023:16 Pages 4317—4330
DOI https://doi.org/10.2147/JIR.S426004
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Zhe Xiong,1,2 Ying Fang,1,2 Shuangshuang Lu,1 Qiuyue Sun,1,3 Jin Huang1
1Department of Gastroenterology, the Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou, Jiangsu Province, People’s Republic of China; 2Graduate School of Dalian Medical University, Dalian, Liaoning Province, People’s Republic of China; 3Graduate School of Nanjing Medical University, Nanjing, Jiangsu Province, People’s Republic of China
Correspondence: Jin Huang, Email [email protected]
Background: Inflammatory bowel disease (IBD) and periodontitis (PD) are correlated, although the pathogenic mechanism behind their correlation has not been clarified. This study aims to explore the common signature genes and potential therapeutic targets of IBD and PD using transcriptomic analysis.
Methods: The GEO database was used to download datasets of IBD and PD, and differential expression analysis was used to identify DEGs. We then conducted GO and KEGG enrichment analyses of the shared genes. Next, we applied 4 machine learning (ML) algorithms (GLM, RF, GBM, and SVM) to select the best prediction model for diagnosing the disease and obtained the hub genes of IBD and PD. The diagnostic value of the signature genes was verified by a validation set and qRT‒PCR experiments. Subsequently, immune cell infiltration in IBD samples and PD samples was analyzed by ssGSEA. Finally, we investigated and validated the response of hub genes to infliximab therapy.
Results: We identified 43 upregulated genes as shared genes by intersecting the DEGs of IBD and PD. Functional enrichment analysis suggested that the shared genes were closely associated with immunity and inflammation. The ML algorithm and qRT‒PCR results indicated that IGKC and COL4A1 were the hub genes with the most diagnostic value for IBD and PD. Subsequently, through immune infiltration analysis, CD4 T cells, NK cells and neutrophils were identified to play crucial roles in the pathogenesis of IBD and PD. Finally, through in vivo and in vitro experiments, we found that IGKC and COL4A1 were significantly downregulated during the treatment of patients with IBD using infliximab.
Conclusion: We investigated the potential association between IBD and PD using transcriptomic analysis. The IGKC and COL4A1 genes were identified as characteristic genes and novel intervention targets for these two diseases. Infliximab may be used to treat or prevent IBD and PD.
Keywords: Inflammatory bowel disease, periodontitis, machine learning, ssGSEA, infliximab, IGKC, COL4A1
Introduction
Inflammatory bowel disease (IBD) is a chronic recurrent autoimmune-related inflammatory disease that includes ulcerative colitis (UC) and Crohn’s disease (CD). At present, the pathophysiology of IBD is still not completely known, but it mainly involves genetic susceptibility, gut microbiota, lifestyle, environmental factors and immune inflammatory responses.1,2 In clinical practice, in addition to gastrointestinal symptoms, approximately 1/3 of IBD patients suffer extraintestinal manifestations (EIMs), which mostly affect the joints, skin, eyes and biliary system.3 Additionally, the oral cavity is also the most common site of EIM involvement, especially in patients with CD.4 Periodontitis (PD) is a chronic inflammatory disease with a pathogenesis similar to that of IBD. It mainly includes the interaction of dysbiosis of the oral microbiota and the immune inflammatory response, eventually leading to periodontal tissue damage and tooth loss.5,6 Currently, several studies have shown that PD is related to various chronic diseases, including atherosclerosis, type 2 diabetes, nonalcoholic fatty liver disease, ulcerative colitis and Crohn’s disease.7–9 Therefore, we need to explore the potential association between IBD and PD, which has important clinical implications for reducing and preventing disease.
In recent years, many epidemiological studies have indicated that the incidence and severity of PD is higher in IBD patients than in healthy control individuals.10,11 In addition, a Mendelian randomization study also suggested that IBD was associated with an increased risk of PD.12 In a previous study, Pietropaoli’s team demonstrated a positive correlation between the severity of PD and the severity of intestinal inflammation by establishing a SAMP1/YitFc (SAMP) mouse model of Crohn’s disease.13 Notably, according to microbiome analysis, studies have revealed a considerable increase in gut colonization by oral bacteria in patients with IBD compared to the healthy population.14 This suggested that oral microbial disorders also influence the progression of bowel inflammation. In several animal studies, oral administration of pathogenic bacteria of PD (Porphyromonas gingivalis, Fusobacterium nucleatum and Haemophilus parainfluenzae) induced intestinal microbiota imbalance and barrier function damage in mice, thus causing a gut inflammation response.15–17 Similarly, PD was also involved in the development of colon inflammation by the swallowing of salivary microbiota to the intestine.18 Additionally, the number of pathogenic Th17 cells (T helper 17) in the oral cavity were increased in patients with PD, and these cells could migrate from the oral cavity to the gut, eventually contributing to gut inflammation.19 However, genetic associations between IBD and PD have rarely been investigated. Therefore, further exploration of the signature genes and molecular mechanisms of IBD and PD could assist in the early diagnosis of both diseases in the future.
Infliximab (IFX), as a representative antitumor necrosis factor (anti-TNF) drug, is a common therapeutic regimen for the treatment of patients with refractory IBD in clinical practice, and it can effectively alleviate the clinical symptoms of the disease to improve the quality of life of patients.20 However, there are studies suggesting that approximately 20% of patients with IBD show nonresponse to IFX therapy.21 Therefore, we explored the response of signature genes of IBD and PD to IFX therapy to provide options for preventing the occurrence of these two diseases and potential targets for the treatment of both diseases.
To understand early diagnostic and therapeutic targets for IBD and PD, we obtained differentially expressed genes for IBD and PD using microarrays. Then, four machine learning (ML) models were utilized to screen for signature genes between the two diseases. Then, to further understand the molecular mechanism underlying the relationship between IBD and PD, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed. Subsequently, we illustrated immune cell infiltration in IBD and PD samples and calculated the correlation between signature genes and immunity. Finally, we investigated the response of the signature genes to infliximab treatment to provide treatment and prevention options for IBD and PD.
Materials and Methods
Dataset Sources
The IBD and PD datasets were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nih.gov/geo/). The GSE7521422 and GSE5907123 datasets were used for the GPL6244 platform. The GSE75214 dataset included 74 samples of active ulcerative colitis, 59 samples of active Crohn’s disease and 22 healthy control samples. GSE59071 included 74 samples of active ulcerative colitis, 8 samples of active Crohn’s disease and 11 healthy control samples. The GSE1613424 and GSE1033425 datasets were used on the GPL570 platform. The GSE16134 dataset included 241 samples with PD and 69 healthy control samples, and the GSE10334 included 183 samples with PD and 64 healthy control samples. Table 1 shows more information on these datasets. Figure 1 illustrates the analysis process of this study.
 |
Table 1 Information on the GEO Datasets |
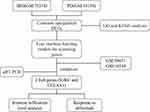 |
Figure 1 Flowchart of this study. |
Identification of Differentially Expressed Genes
Using the limma package,26 we explored the GSE75214 and GSE16134 datasets for differentially expressed genes (DEGs). The selection criteria for GSE75214 were adjusted P value < 0.05 and |log FC (fold change) | ≥ 1. The selection criteria for GSE16134 were adjusted P value < 0.05 and |log FC (fold change) | ≥ 1. Finally, R (4.2.2) software was used to construct the heatmap and volcano plot of the DEGs.
Identification and Enrichment Analysis of Shared Genes
The upregulated DEGs for IBD and PD were intersected to obtain shared genes, and the results are presented using a Venn diagram. Next, the GOplot and the clusterprofiler packages27 were used to conduct GO and KEGG enrichment analysis on the shared genes.
Machine Learning Screening for Hub Genes
To further screen for the hub genes of IBD and PD, we applied 4 machine learning algorithms, including the generalized linear model (GLM), random forest (RF), gradient boosting machine (GBM) and support vector machine (SVM), to calculate the importance of shared genes. Next, each model was examined using the DALEX package to obtain the cumulative residual distribution, and boxplots and the area under the curve (AUC) of the receiver operating characteristic (ROC) curve were utilized to obtain the best model.
Validation of Hub Genes
We evaluated the expression levels of signature genes by the ggplot2 package using boxplots (P < 0.05). The diagnostic value of the signature genes in the IBD and PD datasets (GSE75214, GSE16134, GSE59071 and GSE10334) was evaluated by the pROC package using the AUC of ROC.28
ssGSEA
By ssGSEA, this study investigated the infiltration levels of 28 immune cells in disease (IBD and PD) and healthy samples, and the correlation between hub genes and immune cells was evaluated using Spearman’s rank correlation coefficient (P < 0.05).
Response to Infliximab
To understand the response of hub genes to infliximab, the GSE1458029 dataset (containing 8 responders and 16 non-responders to infliximab for IBD) was downloaded from the GEO database to plot boxplots (P < 0.05).
Sample Collection
To collect intestinal tissue samples, 15 patients were recruited from the Gastroenterology Centre of Changzhou Second People’s Hospital Affiliated with Nanjing Medical University, including 5 healthy patients, 5 patients diagnosed with IBD (3 UC patients and 2 CD patients) and 5 UC patients who responded to therapy with infliximab. The gingival tissue specimens were separated into two groups: the control group, comprising gingival tissue from 4 healthy patients who required surgery for crown lengthening, and the PD group, comprising 4 patients who were found to have severe PD in our dental department and required periodontal flap surgery. All gingival tissues collected measured approximately 2 mm3. The study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Medical Ethics Committee of Changzhou Second People’s Hospital Affiliated with Nanjing Medical University. All patients recruited were informed about the study and signed an informed consent form.
RNA Transcription and Quantitative Real-Time PCR
Total RNA was extracted from tissue samples by an RNAiso plus kit (TaKaRa) and then reverse transcribed to cDNA with cDNA synthesis agent (Vazyme R222-01). qRT‒PCR was subsequently performed using SYBR Green Master Mix reagent (Vazyme Q131), and three replicates of each experiment were performed. The relative data were determined using the 2−∆∆t method, and β-actin was used as an internal control. Table 2 shows the primer sequences.
 |
Table 2 The Primer Sequences for PCR |
Result
Identification of DEGs and Shared Genes
In the IBD dataset GSE75214, 658 DEGs were screened; there were 415 upregulated DEGs and 243 downregulated DEGs. A total of 159 DEGs were screened in the PD dataset GSE16134; there were 125 upregulated DEGs and 34 downregulated DEGs. Among these DEGs, the first 15 DEGs are presented as heatmaps (Figure 2A and B) for IBD and PD, and the expression levels of these DEGs for both diseases are presented as volcano maps (Figure 2C and D). A Venn diagram (Figure 2E) indicated 43 overlapping upregulated DEGs between IBD and PD, and these are defined as shared genes.
Enrichment Analysis of Shared Genes
To investigate the potential regulatory pathways of IBD and PD, we conducted GO and KEGG enrichment analyses on the 43 common genes. According to the GO enrichment analysis (Figure 3A), the shared genes were mainly enriched in immune-related activities, such as the activation of the immune response, leukocyte migration and cytokine-mediated signaling pathways. KEGG enrichment analysis (Figure 3B) indicated that the shared genes were mostly associated with lipids and atherosclerosis, rheumatoid arthritis and the IL-17 signaling pathway.
Identification of Hub Genes by Machine Learning
To obtain the hub genes of IBD and PD, the GLM, RF, GBM and SVM machine learning algorithms were used to construct diagnostic models with features. In the diagnostic model for IBD, the four models were then analyzed using the “DALEX” R package, and the residual box line plot (Figure 4A) was plotted. Based on the reverse cumulative distribution of residuals (Figure 4B) and the area under the curve (Figure 4C), RF (AUC = 1.000) and SVM (AUC = 1.000) were selected as the best models for the diagnosis of IBD. The same methods were used to find the hub genes for PD (Figure 4D and E), and SVM (AUC = 0.847) was the best model for the diagnosis of PD (Figure 4F). The top five genes from each model were selected for further screening. Finally, two overlapping genes (IGKC and COL4A1) (Figure 4G) were identified that could serve as hub genes for IBD and PD.
Validation of Hub Genes
Next, we validated the expression levels and diagnostic value of IGKC and COL4A1 in independent test sets (GSE75214 and GSE16134) and validation sets (GSE59071 and GSE10334) for IBD and PD. Figure 5A and B shows that IGKC and COL4A1 were significantly upregulated in both IBDs. In the GSE75214 dataset (Figure 5C), these hub genes had excellent diagnostic value for IBD: IGKC (AUC = 0.931) and COL4A1 (AUC = 0.978). In the GSE16134 dataset (Figure 5D), IGKC (AUC = 0.902) and COL4A1 (AUC = 0.900) had good diagnostic efficacy. Moreover, Figure 5E and F shows that IGKC and COL4A1 were significantly upregulated in PD. IGKC (AUC = 0.955) and COL4A1 (AUC = 0.992) had a high diagnostic value in the GSE59071 validation dataset (Figure 5G). In the GSE10334 validation dataset (Figure 5H), IGKC (AUC = 0.876) and COL4A1 (AUC = 0.866) were also of high diagnostic value.
Immune Cell Infiltration in IBD and PD
Immune and inflammatory responses are crucial in the molecular pathways of IBD and PD. ssGSEA was further applied to analyze the differences in immune cell infiltration between these two diseases and healthy controls. Figure 6A and B shows the distribution of 28 immune cells in the GSE75214 sample. Figure 7A and B shows the distribution of 28 immune cells in the GSE10334 sample. According to the immune infiltration results, we found that B cells, CD4 T cells, NK cells and neutrophils were strongly correlated with both diseases. Furthermore, the correlation analysis results of immune cells with the hub genes (IGKC and COL4A1) suggested that various immune cells had a positive correlation with IGKC and COL4A1 in IBD samples (Figure 6C) and PD samples (Figure 7C), and active B cells, immature B cells, MDSCs, NK cells and dendritic cells were more highly correlated with IGKC and COL4A1.
Response of Hub Genes to Infliximab and in vitro Experiments
We identified the response of IGKC and COL4A1 to infliximab using the GSE14580 dataset. Figure 8A indicates that IGKC and COL4A1 were significantly downregulated in the responsive group. According to the qRT‒PCR verification results of the hub genes shown in Figure 8B and C, in the comparison between the healthy control and IBD groups, IGKC and COL4A1 expression levels were higher in the IBD group. In the comparison between healthy control and PD groups, IGKC and COL4A1 expression levels were significantly upregulated in the PD group. The findings of qRT‒PCR were in line with the transcriptomic analysis. The above results suggest that IGKC and COL4A1 may be important diagnostic markers for the comorbidity of these two diseases. In addition, in vitro trials showed significant downregulation of IGKC and COL4A1 expression in patients who responded to long-term infliximab therapy compared to patients with active IBD.
Discussion
This work utilized bioinformatics to investigate the interactions between genes and molecular pathways in IBD and PD and to further reveal the relevance of these two diseases to immune cells. In this study, by intersecting the upregulated DEGs obtained from the IBD and PD datasets, we identified 43 genes as shared genes between these two diseases and found that these shared genes contribute to the occurrence and advancement of disease mainly through immune and inflammatory pathways. Subsequently, through machine learning models, IGKC and COL4A1 were identified as hub genes for both diseases and were validated by the validation set and laboratory findings. Subsequently, multiple immune cells were significantly expressed in the IBD and PD samples, according to immune infiltration analysis. Finally, drug response analysis suggested that infliximab significantly affected the expression of IGKC and COL4A1 in patients with IBD.
The enrichment analysis in this study suggested that the shared genes between IBD and PD are mainly closely related to immune/inflammation processes, such as the immune response, leukocyte migration, cytokine‒cytokine receptor interactions, cytokine-mediated signaling pathways, and TNF and IL-17 signaling pathways. It is well known that both IBD and PD are chronic inflammatory diseases. Proinflammatory factors are key factors in the pathogenesis of both diseases. In IBD, neutrophil activation damages the barrier function of the intestinal epithelium through proinflammatory factors. As the number of macrophages in the intestinal mucosa increases, the intestinal mucosa produces large amounts of proinflammatory factors and chemokines (including TNF-α, IL-12 and IL-23), thus aggravating intestinal epithelial damage.30 In several models of PD induced by the use of Porphyromonas gingivalis, proinflammatory factors (such as IL-6, IL-17 and IL-23) secreted mainly by Th17 cells were associated with the pathogenesis of PD, with IL-17 promoting alveolar bone destruction by upregulating the expression level of nuclear factor-kb ligand (RANKL).31 In a previous study, Kitamoto’s team found that the introduction of Klebsiella and Enterobacter species into the gut caused by PD could induce colitis by activating IL-1β secretion by macrophages.19 In addition, the pathogenic bacteria of periodontitis, Fusobacterium nucleatum, increases intestinal permeability by disrupting the intestinal epithelial barrier, which induces the production of cytokines (TNF-α, INF-γ, IL-1β, IL-6 and IL-17), thereby exacerbating gut inflammation.32 However, the Figueredo team found elevated expression levels of cytokines (IL-4, IL-10 and IL-21) in the gingival tissue of individuals with active IBD.33 In conclusion, the inflammatory response may be one of the important pathways for the interaction between IBD and PD, in which the signaling pathway mediated by proinflammatory factors plays a key role.
As with the inflammatory response, the immune response is equally involved in the pathogenesis of IBD and PD, so we further explored the relationship between 28 immune cells and these two diseases. Based on the results of ssGSEA, this study found that the IBD and PD groups were substantially related to multiple immune cells when compared with the control group. It has been reported that pathogenic Th17 cells caused by PD could migrate from the oral cavity into the gut and amplify ectopically colonized Klebsiella, thereby exacerbating intestinal inflammation.19 Furthermore, the common causative agent in PD (Porphyromonas gingivalis) stimulates the CD4 T-cell response, increasing the Th17/Treg ratio, increasing the expression of Th17-related transcription factors and the secretion of proinflammatory factors (IL-17 and IL-4) by activating the TLR17/TLR4-mediated signaling pathway, and decreasing the expression of the Treg transcription factor Foxp3 and the secretion of anti-inflammatory factors (TGF-β and IL-10), resulting in increased gut inflammation.34 In a study of IBD, Yuan’s team revealed that long-term antibiotic application resulted in dysbiosis of the intestinal microbiota, which not only disrupted the intestinal barrier but also increased bone resorption in PD by disrupting the Th17/Treg balance. Next, the application of fecal microbial transplantation (FMT) in mice not only improved the disturbance of the gut microbiome but also alleviated PD symptoms by reversing the Th17/Treg imbalance in periodontal tissue.35 All the above studies suggest that Th17 cells are important immune cells in the interaction between IBD and PD, which is similar to our findings from the immune infiltration analysis. Additionally, this study found that B cells, CD4 T cells, NK cells and neutrophils were highly expressed in IBD and PD. This may bring new insights to the study of the pathological mechanisms of IBD and PD.
After crossing the candidate genes in the best machine learning model and qRT‒PCR experiments, IGKC and COL4A1 were found to be the genes with the most diagnostic value for IBD and PD. In a study based on an immunoglobulin assay, IGKC was identified as a diagnostic marker for screening for PD.36 Recently, there have been many studies on the IGKC gene in the field of oncology, but there is a lack of systematic studies in both IBD and PD. In several studies on breast, colon and lung cancer, immunoglobulin kappa chain (IGKC), one of the top B-cell metagenes, is an important immune marker for predicting survival and response to adjuvant chemotherapy.37,38 Collagen type IV (Col IV) is the most abundant component of basement membranes of the extracellular matrix (ECM) and is produced and secreted mainly by fibroblasts,39,40 endothelial cells and epithelial cells. In a study of transcriptomic analysis, the ECM-related gene COL4A1 was significantly expressed in patients with CD.41 This may be related to Crohn’s disease promoting intestinal fibrosis. Similarly, COL4A1 was found to be significantly upregulated in the development of cardiac fibrosis,42 renal fibrosis,43 and pulmonary fibrosis.44 Furthermore, the upregulation of COL4A1 could promote the growth and metastasis of hepatocellular carcinoma by activating the FAK-Src signaling pathway.45 COL4A1 could also be a potential gene for diagnosis, prognosis, recurrence and trastuzumab resistance in gastric cance.46–48 In the present study, we found that IGKC was closely related to B cells and MDSCs in IBD samples and PD samples, which may indicate that IGKC is a B-cell metagene. Similarly, COL4A1 was involved in the immune response. In neoplastic samples, COL4A1 was positively correlated mainly with macrophages, dendritic cells, B cells and neutrophils.49 According to the immune infiltration results, COL4A1 was positively correlated with many immune cells in IBD samples and PD samples; it was mainly correlated with B cells, dendritic cells and MDSCs. The above findings suggest that IGKC and COL4A1 are involved in the pathological mechanisms of IBD and PD through the regulation of immune cells.
At present, biologics provide a novel approach for the management of patients with refractory IBD. For example, infliximab, an anti-tumor necrosis factor (anti-TNF) biologic,20 effectively improves clinical symptoms and intestinal mucosal healing in patients with IBD.50 However, there are still some patients with IBD who do not respond to biologic therapy in clinical practice.21 Therefore, we then explored whether the hub genes (IGKC and COL4A1) of IBD and PD responded to infliximab therapy. Through relevant datasets and in vitro experiments, we found that IGKC and COL4A1 were significantly downregulated in the infliximab response group. This suggests that infliximab may reduce the risk of PD in patients with IBD and that IGKC and COL4A1 are potential genes that respond to infliximab.
The present study has several limitations. Our findings and laboratory studies rely on a dataset of patients with IBD or PD rather than patients with both diseases. Therefore, in the future, we need to establish animal models of the combination of IBD and PD to validate the potential connection between these two diseases, including the key pathways and immune cells.
Conclusion
This study is the first to reveal the genes interactions and potential mechanisms of IBD and PD using transcriptomic analysis. The IGKC and COL4A1 genes were identified as the most significant common signature genes of IBD and PD, which presents potential targets for the therapy of both diseases. Immune and inflammatory responses may be a common pathological mechanism of these two diseases. For further drug response analysis, infliximab may be used to treat or prevent IBD and PD.
Acknowledgments
This work was supported by the Fund of Nanjing Medical University (NMUC2021026A).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kobayashi T, Siegmund B, Le Berre C, et al. Ulcerative colitis. Nat Rev Dis Primers. 2020;6(1):74. doi:10.1038/s41572-020-0205-x
2. Roda G, Chien Ng S, Kotze PG, et al. Crohn’s disease. Nat Rev Dis Primers. 2020;6(1):22. doi:10.1038/s41572-020-0156-2
3. Vavricka SR, Schoepfer A, Scharl M, Lakatos PL, Navarini A, Rogler G. Extraintestinal Manifestations of Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21(8):1982–1992. doi:10.1097/MIB.0000000000000392
4. Lankarani KB, Sivandzadeh GR, Hassanpour S. Oral manifestation in inflammatory bowel disease: a review. World J Gastroenterol. 2013;19(46):8571–8579. doi:10.3748/wjg.v19.i46.8571
5. Graves D. Cytokines that promote periodontal tissue destruction. J Periodontol. 2008;79(8 Suppl):1585–1591. doi:10.1902/jop.2008.080183
6. Madsen GR, Bertl K, Pandis N, Stavropoulos A, Burisch J. The Impact of Periodontitis on Inflammatory Bowel Disease Activity. Inflamm Bowel Dis. 2023;29(3):396–404. doi:10.1093/ibd/izac090
7. Genco RJ, Borgnakke WS. Risk factors for periodontal disease. Periodontol 2000. 2013;62(1):59–94. doi:10.1111/j.1600-0757.2012.00457.x
8. Zeng Y, Cao S, Chen M. Integrated analysis and exploration of potential shared gene signatures between carotid atherosclerosis and periodontitis. BMC Med Genomics. 2022;15(1):227. doi:10.1186/s12920-022-01373-y
9. Zhang Y, Qiao D, Chen R, Zhu F, Gong J, Yan F. The Association between Periodontitis and Inflammatory Bowel Disease: a Systematic Review and Meta-analysis. Biomed Res Int. 2021;2021:6692420. doi:10.1155/2021/6692420
10. Bertl K, Burisch J, Pandis N, Bruckmann C, Klinge B, Stavropoulos A. Periodontitis prevalence in patients with ulcerative colitis and Crohn’s disease - PPCC: a case-control study. J Clin Periodontol. 2022;49(12):1262–1274. doi:10.1111/jcpe.13615
11. Vavricka SR, Manser CN, Hediger S, et al. Periodontitis and gingivitis in inflammatory bowel disease: a case-control study. Inflamm Bowel Dis. 2013;19(13):2768–2777. doi:10.1097/01.MIB.0000438356.84263.3b
12. Wang Z, Li S, Tan D, et al. Association between inflammatory bowel disease and periodontitis: a bidirectional two-sample Mendelian randomization study. J Clin Periodontol. 2023;50(6):736–743. doi:10.1111/jcpe.13782
13. Pietropaoli D, Del Pinto R, Corridoni D, et al. Occurrence of spontaneous periodontal disease in the SAMP1/YitFc murine model of Crohn disease. J Periodontol. 2014;85(12):1799–1805. doi:10.1902/jop.2014.140316
14. Imai J, Ichikawa H, Kitamoto S, et al. A potential pathogenic association between periodontal disease and Crohn’s disease. JCI Insight. 2021;6(23). doi:10.1172/jci.insight.148543
15. Liu Y, Huang W, Dai K, et al. Inflammatory response of gut, spleen, and liver in mice induced by orally administered Porphyromonas gingivalis. J Oral Microbiol. 2022;14(1):2088936. doi:10.1080/20002297.2022.2088936
16. Settem RP, El-Hassan AT, Honma K, Stafford GP, Sharma A. Fusobacterium nucleatum and Tannerella forsythia induce synergistic alveolar bone loss in a mouse periodontitis model. Infect Immun. 2012;80(7):2436–2443. doi:10.1128/IAI.06276-11
17. Sohn J, Li L, Zhang L, et al. Periodontal disease is associated with increased gut colonization of pathogenic Haemophilusparainfluenzae in patients with Crohn’s disease. Cell Rep. 2023;42(2):112120. doi:10.1016/j.celrep.2023.112120
18. Qian J, Lu J, Huang Y, et al. Periodontitis Salivary Microbiota Worsens Colitis. J Dent Res. 2022;101(5):559–568. doi:10.1177/00220345211049781
19. Kitamoto S, Nagao-Kitamoto H, Jiao Y, et al. The Intermucosal Connection between the Mouth and Gut in Commensal Pathobiont-Driven Colitis. Cell. 2020;182(2):447–462.e414. doi:10.1016/j.cell.2020.05.048
20. Rutgeerts P, Sandborn WJ, Feagan BG, et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353(23):2462–2476. doi:10.1056/NEJMoa050516
21. Wong U, Cross RK. Primary and secondary nonresponse to infliximab: mechanisms and countermeasures. Expert Opin Drug Metab Toxicol. 2017;13(10):1039–1046. doi:10.1080/17425255.2017.1377180
22. Vancamelbeke M, Vanuytsel T, Farré R, et al. Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease. Inflamm Bowel Dis. 2017;23(10):1718–1729. doi:10.1097/MIB.0000000000001246
23. Vanhove W, Peeters PM, Staelens D, et al. Strong Upregulation of AIM2 and IFI16 Inflammasomes in the Mucosa of Patients with Active Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21(11):2673–2682. doi:10.1097/MIB.0000000000000535
24. Papapanou PN, Behle JH, Kebschull M, et al. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiol. 2009;9:221. doi:10.1186/1471-2180-9-221
25. Demmer RT, Behle JH, Wolf DL, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79(11):2112–2124. doi:10.1902/jop.2008.080139
26. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
27. Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi:10.1093/bioinformatics/btv300
28. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12(1):77. doi:10.1186/1471-2105-12-77
29. Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58(12):1612–1619. doi:10.1136/gut.2009.178665
30. Bain CC, Schridde A. Origin, Differentiation, and Function of Intestinal Macrophages. Front Immunol. 2018;9:2733. doi:10.3389/fimmu.2018.02733
31. Cheng WC, Hughes FJ, Taams LS. The presence, function and regulation of IL-17 and Th17 cells in periodontitis. J Clin Periodontol. 2014;41(6):541–549. doi:10.1111/jcpe.12238
32. Liu H, Hong XL, Sun TT, Huang XW, Wang JL, Xiong H. Fusobacterium nucleatum exacerbates colitis by damaging epithelial barriers and inducing aberrant inflammation. J Dig Dis. 2020;21(7):385–398. doi:10.1111/1751-2980.12909
33. Figueredo CM, Martins AP, Lira-Junior R, et al. Activity of inflammatory bowel disease influences the expression of cytokines in gingival tissue. Cytokine. 2017;95:1–6. doi:10.1016/j.cyto.2017.01.016
34. Jia L, Wu R, Han N, et al. Porphyromonas gingivalis and Lactobacillus rhamnosus GG regulate the Th17/Treg balance in colitis via TLR4 and TLR2. Clin Transl Immunol. 2020;9(11):e1213. doi:10.1002/cti2.1213
35. Yuan X, Zhou F, Wang H, et al. Systemic antibiotics increase microbiota pathogenicity and oral bone loss. Int J Oral Sci. 2023;15(1):4. doi:10.1038/s41368-022-00212-1
36. Kerishnan JP, Mohammad S, Alias MS, et al. Identification of biomarkers for periodontal disease using the immunoproteomics approach. PeerJ. 2016;4:e2327. doi:10.7717/peerj.2327
37. Schmidt M, Hellwig B, Hammad S, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin κ C as a compatible prognostic marker in human solid tumors. Clin Cancer Res. 2012;18(9):2695–2703. doi:10.1158/1078-0432.CCR-11-2210
38. Schmidt M, Micke P, Gehrmann M, Hengstler JG. Immunoglobulin kappa chain as an immunologic biomarker of prognosis and chemotherapy response in solid tumors. Oncoimmunology. 2012;1(7):1156–1158. doi:10.4161/onci.21653
39. Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3(6):422–433. doi:10.1038/nrc1094
40. Karsdal MA, Nielsen SH, Leeming DJ, et al. The good and the bad collagens of fibrosis - Their role in signaling and organ function. Adv Drug Deliv Rev. 2017;121:43–56. doi:10.1016/j.addr.2017.07.014
41. Eshelman MA, Harris L, Deiling S, Koltun WA, Jeganathan NA, Yochum GS. Transcriptomic analysis of ileal tissue from Crohn’s disease patients identifies extracellular matrix genes that distinguish individuals by age at diagnosis. Physiol Genomics. 2020;52(10):478–484. doi:10.1152/physiolgenomics.00062.2020
42. Chen S, Puthanveetil P, Feng B, Matkovich SJ, Dorn GW, Chakrabarti S. Cardiac miR-133a overexpression prevents early cardiac fibrosis in diabetes. J Cell Mol Med. 2014;18(3):415–421. doi:10.1111/jcmm.12218
43. Solé C, Moliné T, Vidal M, Ordi-Ros J, Cortés-Hernández J. An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis. Cells. 2019;8(8):773. doi:10.3390/cells8080773
44. Savin IA, Markov AV, Zenkova MA, Sen’kova AV. Asthma and Post-Asthmatic Fibrosis: a Search for New Promising Molecular Markers of Transition from Acute Inflammation to Pulmonary Fibrosis. Biomedicines. 2022;10(5):1017. doi:10.3390/biomedicines10051017
45. Wang T, Jin H, Hu J, et al. COL4A1 promotes the growth and metastasis of hepatocellular carcinoma cells by activating FAK-Src signaling. J Exp Clin Cancer Res. 2020;39(1):148. doi:10.1186/s13046-020-01650-7
46. Huang R, Gu W, Sun B, Gao L. Identification of COL4A1 as a potential gene conferring trastuzumab resistance in gastric cancer based on bioinformatics analysis. Mol Med Rep. 2018;17(5):6387–6396. doi:10.3892/mmr.2018.8664
47. Li T, Gao X, Han L, Yu J, Li H. Identification of hub genes with prognostic values in gastric cancer by bioinformatics analysis. World J Surg Oncol. 2018;16(1):114. doi:10.1186/s12957-018-1409-3
48. Li F, Wang NN, Chang X, et al. Bioinformatics analysis suggests that COL4A1 may play an important role in gastric carcinoma recurrence. J Dig Dis. 2019;20(8):391–400. doi:10.1111/1751-2980.12758
49. Yang J, Hao R, Zhang Y, Deng H, Teng W, Wang Z. Construction of circRNA-miRNA-mRNA network and identification of novel potential biomarkers for non-small cell lung cancer. Cancer Cell Int. 2021;21(1):611. doi:10.1186/s12935-021-02278-z
50. Ben-Horin S, Kopylov U, Chowers Y. Optimizing anti-TNF treatments in inflammatory bowel disease. Autoimmun Rev. 2014;13(1):24–30. doi:10.1016/j.autrev.2013.06.002
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







