Back to Journals » Infection and Drug Resistance » Volume 15
Identification and Genetic Characterization of Fasciola hepatica Isolated from Cattle in Jeddah, Saudi Arabia Based on Sequence Analysis of Mitochondrial (COI) Gene
Authors Alsulami MN, Wakid MH , Al-matary M, Abdel-Gaber R , Al-Megrin WAI, Bakhraibah AO, Alanazi AD, Elshabrawy HA, El-Kady AM
Received 21 May 2022
Accepted for publication 29 July 2022
Published 26 August 2022 Volume 2022:15 Pages 4877—4886
DOI https://doi.org/10.2147/IDR.S375671
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Muslimah N Alsulami,1 Majed H Wakid,2 Mohammed Al-matary,3 Rewaida Abdel-Gaber,4 Wafa Abdullah I Al-Megrin,5 Areej O Bakhraibah,1 Abdullah D Alanazi,6 Hatem A Elshabrawy,7 Asmaa M El-Kady8
1Department of Biology, College of Science, University of Jeddah, Jeddah, 21493, Saudi Arabia; 2Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, 21589, Saudi Arabia; 3Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, 21589, Saudi Arabia; 4Department of Zoology, College of Science, King Saud University, Riyadh, 11451, Saudi Arabia; 5Department of Biology, College of Science, Princess Nourah bint Abdulrahman University, Riyadh, 11671, Saudi Arabia; 6Department of Biological Sciences, Faculty of Science and Humanities, Shaqra University, Ad-Dawadimi, 11911, Saudi Arabia; 7Department of Molecular and Cellular Biology, College of Osteopathic Medicine, Sam Houston State University, Conroe, TX, 77304, USA; 8Department of Medical Parasitology, Faculty of Medicine, South Valley University, Qena, 83523, Egypt
Correspondence: Asmaa M El-Kady; Hatem A Elshabrawy, Email [email protected]; [email protected]
Background: In Saudi Arabia, more than US$ 0.2 million annual losses are caused by liver condemnations due to fascioliasis. Data obtained from the genetic characterization of Fasciola population sheds light on parasite transmission which could eventually help in development of effective parasite control measures. So, the aim of this study was to investigate the genetic diversity of Fasciola spp. isolated from cattle in Saudi Arabia by sequence analyses of COI gene.
Materials and Methods: A total of 325 cows slaughtered at the central municipal abattoir in Jeddah city, Jeddah Province, Saudi Arabia were examined for fascioliasis in the period from 1st of June to 1st of July 2020. DNA was extracted from adult Fasciola worms and used for PCR and DNA sequence using a primer pair targeting COI gene. Analysis of the obtained sequences was done using BLAST search and phylogenetic analysis.
Results: Bovine fascioliasis was diagnosed in 18 out of 325 cattle (5.5%). Forty-eight flukes were extracted from infected animals and DNA was successfully amplified from all flukes. Overall 12 different DNA sequences were obtained. BLAST search showed that all obtained sequences were F. hepatica and had > 97% similarity with F. hepatica isolates from Tanzania, Europe and Iran. Phylogenetic analysis of the obtained sequences showed that Fasciola isolates from the current study were clustered in one subclade closely related to isolates from North and South Africa and Italy.
Conclusion: Reports on the molecular characterization of Fasciola spp. in Saudi Arabia are limited. In the current study, our findings showed that F. hepatica was the only Fasciola species parasitizing cattle in Jeddah city, Saudi Arabia. Further studies using a large number of samples from different localities in Saudi Arabia are needed to provide data that will help the development of control measures against fascioliasis.
Keywords: fascioliasis, cattle, PCR, COI, Saudi Arabia
Introduction
Fascioliasis is a zoonotic helminthic infection caused by Fasciola hepatica and F. gigantica.1 Fascioliasis causes serious economic and health problems; especially in developing countries, due to high cost of anthelmintic drugs.2
The definitive host acquire fascioliasis by ingestion of metacercariae. The disease usually results in decreased animal production of meat, milk, and wool as well as a higher prevalence of secondary bacterial infections, with an annual economic loss of more than 2000 million dollars worldwide.3 Triclabendazole has long been used as the first line of treatment for fascioliasis, due to its therapeutic efficacy against all developmental stages of the parasite.4 However, emerging resistance has been reported to this drug, which complicates the treatment of fascioliasis in farmed ruminants.4
Although, fascioliasis is primarily a disease of ruminants (sheep and cattle), outbreaks of human infections have been reported in the last three decades.5 It was estimated that 118 million humans are infected while 180 million are at risk.6 In the past, the disease was restricted to specific geographical areas; however, according to the World Health Organization (WHO), human cases are increasingly reported in Europe, North and South America, Oceania, Africa and Asia.7
In Saudi Arabia, animal fascioliasis has been reported with prevalence of 13.5% and 52.9% in sheep and cattle respectively.8–10 Fascioliasis is considered the main cause of total liver condemnation in 52.06% of cattle in Saudi Arabia, whereas meat and offal’s condemnation resulted in economic loss of 0.2 million dollars annually.8,9 Moreover, human cases of fascioliasis were reported among immigrant workers in Saudi Arabia.8,11
In epidemiological studies, it is usually difficult to differentiate accurately between Fasciola species based on morphological criteria.12 So, following morphological identification, molecular approaches have been applied for the genotyping of the Fasciola parasite such as random amplified polymorphic DNA (RAPD),12 single nucleotide polymorphism (SNP),13 polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)14 and sequencing of the whole genome.15,16
Data obtained from the genetic characterization of Fasciola population sheds light on the distribution and spread of infections among animals which could eventually help in development of effective parasite control measures and parasite elimination.15,17 Moreover, genetic studies are critical to find the source, infectivity, pathogenicity, evolution and development of anthelmintic drug resistance in parasites.18
Although significant DNA sequencing for F. hepatica and F. gigantica are available from several regions, there is few data for the genotyping of Fasciola spp. in Saudi Arabia. Thus, in the present study, we aimed – for the first time – to genetically characterize Fasciola spp. isolated from cattle by sequence analyses of the mitochondrial cytochrome oxidase subunit 1 gene (COI).
Materials and Methods
Study Area
The current research was conducted in Jeddah city, Jeddah Province, Saudi Arabia, between 1st of June and 1st of July 2020. Jeddah city is a Saudi city located on the eastern bank of the Red Sea. In contrast to other Saudi Arabian cities, Jeddah has a warm winter climate, with temperatures ranging from 15°C in the morning to 28°C in the afternoon. Summers are quite hot and humid, specifically in September. The rainfall in Jeddah is short, with the majority of it falling in November and December (Figure 1).19
 |
Figure 1 A map showing jeddah province in Saudi Arabia (the study area). Redrawn from https://www.wikiwand.com/en/List_of_cities_and_towns_in_Saudi_Arabia.19 |
Parasites Collection
A total of 325 cows, slaughtered in Jeddah Municipal abattoir, were divided into two groups (young <1 year and adults >1 year old) and included in the present study. The liver and gall bladders of slaughtered cows were examined for the presence of Fasciola spp. adult worms. Identification of the collected parasites was done using the dissecting microscope on basis of their morphological features.20 All live worms were collected and incubated in warm RPMI 1640 media (Gibco, Life Technologies, CA, USA; Catalog number: 11875093) for 5 h at 37°C to allow regurgitation to prevent contamination by host blood present in the worm gut. The media was changed 3–5 times during incubation. Collected worms were then washed with saline and transferred into sterile containers with 70% ethanol and refrigerated at 4°C for later use.21
DNA Extraction
A small piece of the apical zone of the posterior end of adult worms (n = 48 isolated from 18 infected cows; two to four worms from each cow) was cut to remove likely contamination by sperm or eggs present in the reproductive organs and was used for DNA extraction.22 This piece was then homogenized using a micro-electric tissue homogenizer and genomic DNA was obtained using the DNeasy Blood & Tissue DNA extraction kit (Qiagen, Germantown, MD, USA, Cat. No./ID: 69504), based on the guidelines of producer.
Polymerase Chain Reaction (PCR)
PCR reaction, in a total volume of 50 µL, was performed using thermal cycler (Z316091 Eppendorf® Mastercycler Personal, AC/DC input 230 V AC, 50–60 Hz) to amplify the partial COI gene. Two primers, Ita8 (forward: 5′-ACGTTGGATCATAAGCGTGT-3′) and Ita9 (reverse: 5′-CCTCATCCAACATAACCTCT-3′) were used according to previous studies.23 The PCR conditions were set as following: initial denaturation at 95°C for 5 min, 36 cycles of denaturation at 95°C for 40 seconds, annealing at 46°C for 55 seconds and extension at 72°C for 1 min. Final extension was done for 10 min at 72°C. The predictable size of the PCR product was 493 bp.
PCR products were analyzed by electrophoresis on a 1% agarose gel (Bioline, Cat. No. BIO-41027) in TAE buffer containing SYBR Safe (Invitrogen, SYBR Safe™, Cat. No. S33102). Electrophoresis was performed at 90V for 90 min. PCR products size was reported by using a GelDoc EZ Imager (Bio-Rad, GelDoc EZ Imager, Cat. No. 1708270).
DNA Sequencing
PCR products (48 adult worms extracted from 18 infected cows) were purified using the Bioline Isolate PCR and Gel kit (London, UK, Isolate II PCR and Gel kit Cat. No. BIO-52059) following the manufacturer’s instructions. PCR products were then sequenced using the same forward and reverse primers used for PCR.
Sequence and Phylogenetic Analyses
Obtained sequences of PCR products were aligned with DNA sequences of Fasciola spp. deposited from different countries in the GenBank using BLAST software. Multiple alignments were done using MUSCLE program of MEGA X software.
Phylogenetic analysis and Pairwise nucleotide variations of COI gene were performed using the Maximum Likelihood method and Tamura 3 parameter model of the MEGA X software. Sequences used for construction of the phylogenetic tree are listed in Table 1.
 |
Table 1 Shows the Sequences Used for Construction of the Phylogenetic Tree |
Statistical Analysis
All obtained data and results were statistically analyzed by SPSS program (version 22). Differences between groups were determined using Chi-square. P values < 0.05 were considered statistically significant.
Ethical Statement
The present study was approved by the Institutional Review Board, College of Science, University of Jeddah, Saudi Arabia (protocol code UJ212430061).
Results
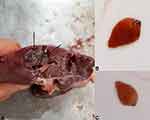
Examination of the livers and gall bladders of 325 cows slaughtered at the central Municipal abattoir in Jeddah city, revealed that 18 cows (5.5%) were infected with Fasciola adult worms (Figure 2). Higher infection rates were reported among male animals (6.9% in males vs 4.0% in females). However, the reported difference was not statistically significant (P=0.9). Similarly, higher infection rates were demonstrated in the adult animal group in comparison to young animals with no statistical significance (P=0.4) (Table 2).
 |
Table 2 Prevalence of F. hepatica in Cows at Jeddah Municipal Abattoir |
Fasciola sp. adult worms (n=48) were collected from the infected cows (n=18). DNA was isolated and successfully amplified from all worms resulting in PCR products of the right size (493 bp) (Figure 3).
Next, we sequenced all PCR products (n = 48) and obtained 12 different DNA sequences. Frequency of sequences is shown in Table 3.
 |
Table 3 Showing the Frequency of 12 Sequences Obtained from 48 Adult Worms Isolated from 18 Infected Cows |
The variance of the obtained sequences ranged from 0.1887 to 1.0185. Detailed results of nucleotide variation are shown in Table 4.
 |
Table 4 Genetic p-Distances Among F. hepatica Isolates in the Present Study Based on Sequence Analysis of Partial COI Gene |
Alignment of the sequences obtained from the current study identified the isolated worms as F. hepatica based on >97% similarity with F. hepatica sequences deposited in the GenBank database. A BLAST search showed that the sequences are similar to F. hepatica isolated from cattle in Tanzania (JX236645 and EU282862), Eastern Europe (HM487172), Netherlands (FJ936028) and Iran (KY246450 and KY246451).
To construct the phylogenetic tree, the Maximum Likelihood method and Tamura-3 model were used. All sequences in the present study were compared with other sequences of F. hepatica deposited in the GenBank using P. westermani as an outgroup (Figure 4). Our isolates clustered with other F. hepatica sequences from the GenBank database which supported BLAST search results and confirmed that all of our isolates were F. hepatica. Moreover, isolates from the present study were clustered in one group close to isolates from North Africa, South Africa, Turkey and Italy.
Discussion
Bovine fascioliasis is often presented as a subclinical disease that decreases animal productivity and results in significant economic loss. Several studies have demonstrated that the control of bovine fascioliasis is complicated by the complexity of the parasite’s life cycle and emerging drug resistance.24 Therefore, understanding the parasite life cycle, and evaluation of its genetic diversity and population structure would allow the development of effective control measures.15,17,24
In the present study, we aimed to identify the prevalence and phylogenetic analysis of fascioliasis in infected cattle for the first time in Jeddah city, Saudi Arabia. A total of 325 slaughtered cattle were examined. Eighteen cows (5.5%) were found to be infected with adult F. hepatica worms. Previous reports from several Saudi Arabian localities showed a wide range of infection rate. In Riyadh, Magda and Al-Megrin reported a higher infection rate in 2005 (21.9%) and Degheidy et al in 2012 (16.9%).8,10 Mgzoub and Kasim, on the other hand, reported a lower infection rate in sheep from different regions in Saudi Arabia.25 The authors reported an infection rate ranged from 0.18% to 2.4%.25 Variable findings from different localities may be explained by the difference in sample size, environmental conditions, and the application of control measures.
To the best of our knowledge, this is the first study in Saudi Arabia to genetically characterize Fasciola sp. in cattle based on COI gene. Based on DNA sequence analysis, the isolated Fasciola adult worms in the present study were identified as F. hepatica. This finding is consistent with previous reports from Saudi Arabia.26 Alajmi, reported F. hepatica as the predominant Fasciola species isolated from sheep in Riyadh, Saudi Arabia. The author identified both F. hepatica and F. gigantica in examined sheep, with F. hepatica inhabiting 80% of examined animals.26 On the other hand, Shalaby et al reported equal infection rates for both F. hepatica and F. gigantica in imported sheep in Al Taif region in Saudi Arabia had.27 Similar results have been reported by Farjallah et al.28 The authors confirmed that all samples from Tunisia and Algeria samples belong to a single species, namely F. hepatica.28 Based on both nuclear and mitochondrial sequences, reports from different worldwide locations had shown that F. hepatica has been identified as the most common Fasciola spp. in temperate regions while F. gigantica is the commonest in tropical countries of Africa.20,29–32
Sequence analysis of the PCR products of COI gene obtained from 48 F. hepatica worms isolated from 18 infected cows in the present study identified 12 different isolates. The phylogenetic analysis of the obtained isolates using MEGA X software showed a rooted tree with Paragonimus westermani as an outgroup. The resulting phylogenetic tree showed a close relationship of our isolates with those of other countries. The obtained isolates (12) clustered in one subclade and were closely related to isolates from Italy (JF824674.1),33 Turkey (GQ121276.1),14 South Africa (KT182261.1),34 Algeria (MK212144.1),35 North Africa (GQ231551.1),28 Tunisia (GQ231550.1)28.
Conclusion
Although there have been multiple publications on fascioliasis in Saudi Arabia, all of these reports are based on the morphology of eggs and adult worms, with limited molecular species identification. In the current study, we used COI as a molecular marker for genetic characterization of Fasciola adult worm isolated from infected cattle. Our findings showed that F. hepatica was the only Fasciola species parasitizing cattle in Jeddah city, Saudi Arabia. The present study provided data for future monitoring of fascioliasis and identifying risk factors. Further studies using new molecular markers will be useful in the control and prevention of fascioliasis.
Study Limitation
Future research with a larger number of Fasciola adult worms from various regions throughout Saudi Arabia in order to figure out prevalence, risk factors, geographical distribution, how it spread, where it is thought to have originated, to other parts of the world, including Saudi Arabia.
Acknowledgments
The authors extend their Acknowledgments to Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R39), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia and the Researchers Supporting Project (RSP-2021/25), King Saud University, Riyadh, Saudi Arabia.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Nyindo M, Lukambagire A-H. Fascioliasis: an ongoing zoonotic trematode infection. Biomed Res Int. 2015;2015:786195. doi:10.1155/2015/786195
2. Daryani A, Alaei R, Arab R, Sharif M, Dehghan MH, Ziaei H. Prevalence of liver fluke infections in slaughtered animals in Ardabil Province, Northwestern Iran. J Anim Vet Adv. 2006;5:3.
3. Mungube EO, Bauni SM, Tenhagen BA, Wamae LW, Nginyi JM, Mugambi JM. The prevalence and economic significance of Fasciola gigantica and Stilesia hepatica in slaughtered animals in the semi-arid coastal Kenya. Trop Anim Health Prod. 2006;38(6):475–483. doi:10.1007/s11250-006-4394-4
4. Kelley JM, Elliott TP, Beddoe T, Anderson G, Skuce P, Spithill TW. Current threat of triclabendazole resistance in Fasciola hepatica. Trends Parasitol. 2016;32(6):458–469. doi:10.1016/j.pt.2016.03.002
5. Haseeb AN, el-Shazly AM, Arafa MAS, ATA M. A review on fascioliasis in Egypt. J Egypt Soc Parasitol. 2002;32(1):317–354.
6. Mas-Coma S, Valero MA, Bargues MD. Chapter 2. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol. 2009;69:41–146. doi:10.1016/S0065-308X(09)69002-3
7. Soliman MFM. Epidemiological review of human and animal fascioliasis in Egypt. J Infect Dev Ctries. 2008;2(3):182–189. doi:10.3855/jidc.260
8. Degheidy NS, Al-Malki JS. Epidemiological studies of fasciolosis in human and animals at Taif, Saudi Arabia. World Appl Sci J. 2012;19(8):1099–1104.
9. Mehmood K, Zhang H, Sabir AJ, et al. A review on epidemiology, global prevalence and economical losses of fasciolosis in ruminants. Microb Pathog. 2017;109:
10. Sanad M, Al-Megrin W. Fascioliasis among local and imported sheep in Saudi Arabia: parasitological and serological diagnosis. J Egypt Soc Parasitol. 2006;35:1121–1134.
11. El-Mathal EM, Fouad MAH. Human fascioliasis among immigrant workers in Saudi Arabia. J Egypt Soc Parasitol. 2005;35(3 Suppl):1199–1207.
12. Aryaeipour M, Rouhani S, Bandehpour M, Mirahmadi H, Kazemi B, Rokni MB. Genotyping and phylogenetic analysis of Fasciola spp. isolated from sheep and cattle using PCR-RFLP in Ardabil Province, Northwestern Iran. Iran J Public Health. 2014;43(10):1364–1371.
13. Cwiklinski K, Dalton JP, Dufresne PJ, et al. The Fasciola hepatica genome: gene duplication and polymorphism reveals adaptation to the host environment and the capacity for rapid evolution. Genome Biol. 2015;16(1):71. doi:10.1186/s13059-015-0632-2
14. Simsek S, Utuk AE, Balkaya I. Molecular differentiation of Turkey cattle isolates of Fasciola hepatica and Fasciola gigantica. Helminthologia. 2011;48(1):3–7. doi:10.2478/s11687-011-0001-y
15. Itagaki T, Kikawa M, Terasaki K, Shibahara T, Fukuda K. Molecular characterization of parthenogenic Fasciola sp. in Korea on the basis of DNA sequences of ribosomal ITS1 and mitochondrial NDI gene. J Vet Med Sci. 2005;67(11):1115–1118. doi:10.1292/jvms.67.1115
16. Marcilla A, Bargues MD, Mas-Coma S. A PCR-RFLP assay for the distinction between Fasciola hepatica and Fasciola gigantica. Mol Cell Probes. 2002;16(5):327–333. doi:10.1006/mcpr.2002.0429
17. Sharifiyazdi H, Moazeni M, Rabbani F. Molecular characterization of human Fasciola samples in Gilan province, Northern Iran on the basis of DNA sequences of ribosomal and mitochondrial DNA genes. Comp Clin Path. 2012;21(5):889–894. doi:10.1007/s00580-011-1193-8
18. Beesley NJ, Williams DJL, Paterson S, Hodgkinson J. Fasciola hepatica demonstrates high levels of genetic diversity, a lack of population structure and high gene flow: possible implications for drug resistance. Int J Parasitol. 2017;47(1):11–20. doi:10.1016/j.ijpara.2016.09.007
19. List of cities and towns in Saudi Arabia, wikiwand [updated 2021]. Available from: https://www.wikiwand.com/en/List_of_cities_and_towns_in_Saudi_Arabia.
20. Valero MA, Panova M, Mas-Coma S. Phenotypic analysis of adults and eggs of Fasciola hepatica by computer image analysis system. J Helminthol. 2005;79(3):217–225. doi:10.1079/JOH2005301
21. Tang M, Zhou Y, Liu Y, Cheng N, Zhang J, Xu X. Molecular identification and genetic-polymorphism analysis of Fasciola flukes in Dali Prefecture, Yunnan Province, China. Parasitol Int. 2021;85:102416. doi:10.1016/j.parint.2021.102416
22. Anh DN, Anh LT, Tuan LQ, et al. Identification of Fasciola species isolates from Nghe An Province, Vietnam, based on ITS1 sequence of ribosomal DNA using a simple PCR-RFLP method. J Parasitol Res. 2018;2018:2958026. doi:10.1155/2018/2958026
23. Itagaki T, Kikawa M, Sakaguchi K, et al. Genetic characterization of parthenogenic Fasciola sp. in Japan on the basis of the sequences of ribosomal and mitochondrial DNA. Parasitology. 2005;131(Pt5):679–685. doi:10.1017/S0031182005008292
24. Kaplan RM. Fasciola hepatica: a review of the economic impact in cattle and considerations for control. Vet Ther. 2001;2(1):40–50.
25. Magzoub M, Kasim AA. The prevalence of fascioliasis in Saudi Arabia. Trop Anim Health Prod. 1978;10(4):205–206. doi:10.1007/BF02235342
26. Alajmi RA. Molecular characterization of Fasciola flukes using mitochondrial 28S rRNA gene in Naimi Saudi sheep. Saudi J Biol Sci. 2019;26(1):112–117. doi:10.1016/j.sjbs.2017.06.010
27. Shalaby I, Gherbawy Y, Banaja A. Molecular characterization of Fasciola species isolated from imported sheep in Taif region (Saudi Arabia). Trop Biomed. 2013;30(1):15–26.
28. Farjallah S, Sanna D, Amor N, et al. Genetic characterization of Fasciola hepatica from Tunisia and Algeria based on mitochondrial and nuclear DNA sequences. Parasitol Res. 2009;105(6):1617–1621. doi:10.1007/s00436-009-1601-z
29. Agatsuma T, Arakawa Y, Iwagami M, et al. Molecular evidence of natural hybridization between Fasciola hepatica and F. gigantica. Parasitol Int. 2000;49(3):231–238. doi:10.1016/s1383-5769(00)00051-9
30. Ai L, Chen M-X, Alasaad S, et al. Genetic characterization, species differentiation and detection of Fasciola spp. by molecular approaches. Parasit Vectors. 2011;4(1):101. doi:10.1186/1756-3305-4-101
31. Raina OK, Jacob SS, Sankar M, et al. Genetic characterization of Fasciola gigantica from different geographical regions of India by ribosomal DNA markers. J Parasit Dis. 2015;39(1):27–32. doi:10.1007/s12639-013-0276-7
32. Walker SM, Makundi AE, Namuba FV, et al. The distribution of Fasciola hepatica and Fasciola gigantica within southern Tanzania–constraints associated with the intermediate host. Parasitology. 2008;135(4):495–503. doi:10.1017/S0031182007004076
33. Farjallah S, Ben Slimane B, Piras CM, Amor N, Garippa G, Merella P. Molecular characterization of Fasciola hepatica from Sardinia based on sequence analysis of genomic and mitochondrial gene markers. Exp Parasitol. 2013;135(3):471–478. doi:10.1016/j.exppara.2013.08.006
34. Mucheka VT, Lamb JM, Pfukenyi DM, Mukaratirwa S. DNA sequence analyses reveal co-occurrence of novel haplotypes of Fasciola gigantica with F. hepatica in South Africa and Zimbabwe. Vet Parasitol. 2015;214(1–2):144–151. doi:10.1016/j.vetpar.2015.09.024
35. Chougar L, Amor N, Farjallah S, et al. New insight into genetic variation and haplotype diversity of Fasciola hepatica from Algeria. Parasitol Res. 2019;118(4):1179–1192. doi:10.1007/s00436-019-06270-5
36. Amer S, Dar Y, Ichikawa M, et al. Identification of Fasciola species isolated from Egypt based on sequence analysis of genomic (ITS1 and ITS2) and mitochondrial (NDI and COI) gene markers. Parasitol Int. 2011;60(1):5–12. doi:10.1016/j.parint.2010.09.003
37. Moazeni M, Sharifiyazdi H, Izadpanah A. Characterization of Fasciola hepatica genotypes from cattle and sheep in Iran using cytochrome C oxidase gene (CO1). Parasitol Res. 2012;110(6):2379–2384. doi:10.1007/s00436-011-2774-9
38. Halakou A, Niloofar T, A Amin, et al. Molecular Identification and phylogenetic analysis of Fasciola hepatica isolates from cattle and sheep in Golestan Province, Northeast of Iran. Res Sq. 2021. doi:10.21203/rs.3.rs-477200/v1
39. Beltrame MO, Pruzzo C, Sanabria R, Pérez A, Mora MS. First report of pre-Hispanic Fasciola hepatica from South America revealed by ancient DNA. Parasitology. 2020;147(3):371–375. doi:10.1017/S0031182019001719
40. van Herwerden L, Blair D, Agatsuma T. Intra- and interindividual variation in ITS1 of paragonimus westermani (Trematoda: digenea) and related species: implications for phylogenetic studies. Mol Phylogenet Evol. 1999;12(1):67–73. doi:10.1006/mpev.1998.0572
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.