Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 19
Identification and Characterization of a ceRNA Regulatory Network Involving LINC00482 and PRRC2B in Peripheral Blood Mononuclear Cells: Implications for COPD Pathogenesis and Diagnosis
Authors Huang W, Luo T, Lan M, Zhou W, Zhang M, Wu L, Lu Z, Fan L
Received 18 October 2023
Accepted for publication 1 February 2024
Published 8 February 2024 Volume 2024:19 Pages 419—430
DOI https://doi.org/10.2147/COPD.S437046
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Wenjie Huang,1,2 Ting Luo,1,2 Mengqiu Lan,3 Wenting Zhou,1,2 Ming Zhang,1,2 Lihong Wu,3 Zhenni Lu,3 Li Fan1,2
1Department of Reproductive Medicine, Guangzhou Women and Children’s Medical Center Liuzhou Hospital, Liuzhou, Guangxi, 545616, People’s Republic of China; 2Department of Reproductive Medicine, Liuzhou Maternity and Child Healthcare Hospital, Liuzhou, Guangxi, 545001, People’s Republic of China; 3Clinical Laboratory Science, Liuzhou Municipal Liutie Central Hospital, Liuzhou, Guangxi, 545007, People’s Republic of China
Correspondence: Wenjie Huang, Email [email protected]
Purpose: Chronic obstructive pulmonary disease (COPD) is the third leading cause of death worldwide, characterized by intense lung infiltrations of immune cells (macrophages and monocytes). While existing studies have highlighted the crucial role of the competitive endogenous RNA (ceRNA) regulatory network in COPD development, the complexity and characteristics of the ceRNA network in monocytes remain unexplored.
Methods: We downloaded messenger RNA (mRNA), microRNA (miRNA), and long noncoding RNA (lncRNA) microarray data from GSE146560, GSE102915, and GSE71220 in the Gene Expression Omnibus (GEO) database. This data was used to identify differentially expressed mRNAs (DEmRNAs), miRNAs (DEmiRNAs), and lncRNAs (DElncRNAs). Predicted miRNAs that bind to DElncRNAs were intersected with DEmiRNAs, forming a set of intersecting miRNAs. This set was then used to predict potential binding mRNAs, intersected with DEmRNAs, and underwent functional enrichment analysis using R software and the STRING database. The resulting triple regulatory network and hub genes were constructed using Cytoscape. Comparative Toxicomics Database (CTD) was utilized for disease correlation predictions, and ROC curve analysis assessed diagnostic accuracy.
Results: Our study identified 5 lncRNAs, 4 miRNAs, and 149 mRNAs as differentially expressed. A lncRNA-miRNA-mRNA regulatory network was constructed, and hub genes were selected through hub analysis. Enrichment analysis highlighted terms related to cell movement and gene expression regulation. We established a LINC00482-has-miR-6088-PRRC2B ceRNA network with diagnostic relevance for COPD. ROC analysis demonstrated the diagnostic value of these genes. Moreover, a positive correlation between LINC00482 and PRRC2B expression was observed in COPD PBMCs. The CTD database indicated their involvement in inflammatory responses.
Conclusion: In summary, our study not only identified pivotal hub genes in peripheral blood mononuclear cells (PBMCs) of COPD but also constructed a ceRNA regulatory network. This contributes to understanding the pathophysiological processes of COPD through bioinformatics analysis, expanding our knowledge of COPD, and providing a foundation for potential diagnostic and therapeutic targets for COPD.
Keywords: COPD, PBMCs, ceRNA, bioinformatics analysis, PRRC2B, LINC00482
Introduction
Chronic obstructive pulmonary disease (COPD) is a significant contributor to global mortality, healthcare burden, and healthcare costs. The mortality rates associated with COPD have seen an alarming increase of 14.1% from 2009 to 2019, attributed to factors such as exposure to inhaled pollutants, tobacco usage, genetic predisposition, and developmental and social influences.1 It’s noteworthy that many individuals who eventually receive a COPD diagnosis experience symptoms as early as midlife, hinting at a protracted disease onset that may begin years before formal diagnosis.2 Additionally, the episodic exacerbations characteristic of COPD, marked by acute deteriorations, lead to substantial negative impacts, including heightened mortality and morbidity following severe and recurrent events.3 However, the heterogeneous nature of these exacerbations and the lack of dependable clinical, physiological, or biological markers pose challenges in predicting the onset of exacerbations.4 Consequently, there’s a growing emphasis on the importance of early COPD detection and ongoing monitoring.
While various biochemical markers have been proposed as predictors of COPD outcomes, their measurement often demands significant time and resources. Clinical trials aiming to predict COPD progression and outcomes through common blood count metrics such as eosinophils, neutrophil/lymphocyte ratio (NLR), platelet/lymphocyte ratio (PLR), and eosinophil/basophil ratio (EBR) have shown promise, but their effectiveness requires validation.5,6 Moreover, the genetic factor alpha-1 antitrypsin deficiency (AATD) is well-established as a contributor to early-onset COPD risk, regardless of smoking history. Studies have even shown that peripheral blood monocytes from COPD patients exhibit different inflammatory cytokine release patterns when comparing individuals with the alpha1-antitrypsin deficiency variant (Pizz) and those with the normal ATT variant (PiMM).7 Circulating monocyte counts, especially the non-classical subset, have been observed to increase significantly in COPD patients, suggesting their potential role in disease progression.8 Furthermore, gene expression signatures derived from peripheral blood mononuclear cells (PBMCs), representative of systemic immune activity, have been shown to play pivotal roles in the pathogenesis of COPD.9 This insight suggests that monitoring monocytes could aid in tracking COPD progression and identifying novel biomarkers.
LncRNAs are a category of non-protein coding RNAs that exert significant influence on various cellular processes, particularly in tumor initiation and progression.10 On the other hand, microRNAs (miRNAs) are short, endogenous non-coding RNAs (~20 nucleotides) known to post-transcriptionally regulate gene expression. The competitive endogenous RNA (ceRNA) hypothesis, introduced by Salmena et al in 2011, posits that lncRNAs function as ceRNAs by competitively binding to miRNAs, thereby modulating their availability for binding with mRNAs.11
While the ceRNA hypothesis has gained extensive validation within the realm of cancer biology, its extension to respiratory diseases such as asthma and COPD has been relatively limited. This study aims to delve into the ceRNA network within peripheral blood mononuclear cells (PBMCs) to unravel its role in the molecular underpinnings of COPD. By doing so, the study aims to shed light on the etiology of COPD, enhance the ability to predict disease risk, and identify potential therapeutic targets.
Materials and Methods
Data Preparation and Preprocessing
Gene expression profiles of peripheral blood monocytes from two datasets were obtained: GSE146560 (8 COPD patients and 8 healthy controls) and GSE102915 (6 COPD patients and 6 healthy controls), sourced from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE71200 was used to evaluate the expression of hub gene. Detailed information of the 3 datasets is provided in Table 1.
 |
Table 1 Details of GEO COPD Data |
Raw data underwent normalization using the “normalizeBetweenArrays” function from the R package “limma”. Platform annotation files were utilized to convert probe expression matrices into gene expression matrices. In cases where different probes corresponded to the same genes, the average value was computed.
Screening of Differentially Expressed Genes
DElncRNAs, DEmiRNAs, and DEmRNAs were identified using specific criteria: |logFC| > 1.5 and p < 0.05 for DElncRNAs and DEmRNAs, and |logFC| > 0.5 and p < 0.05 for DEmiRNAs. Volcano plots for visualizing DEGs were generated using the R package “EnhancedVolcano”. Heatmap clustering of DEGs was constructed using the R package “pheatmap”.
Establishment of the ceRNA Network in COPD
According to the hypothesis that lncRNA could indirectly regulate mRNA expression by competing with miRNA as a natural sponge in the cytoplasm, the ceRNA network was constructed by the following steps: (1) miRNet12 was used to predict the potential miRNAs targeted by DElncRNAs and the lncRNA-miRNA interaction pairs; (2) TargetScan13 and miRDB14 were used to forecast the target genes of the intersection of miRNAs by overlapping the predicted miRNAs with DEmiRNA; (3) The R package “VennDiagram” facilitated the intersection of target genes and DEmRNAs. The resulting intersection of mRNAs was selected for subsequent analysis; (4) Integration of lncRNA-miRNA pairs with miRNA-mRNA pairs resulted in the construction of a lncRNA-miRNA-mRNA triple regulatory network.
The Cytoscape plug-in “CytoHubba” was employed to identify hub triple regulatory networks. The generated networks were visualized using Cytoscape software (version 3.9.1). LncRNA sequences were obtained from LNCipedia,15 and the lncRNA cellular localization was determined using the LncLocator database.16
Functional Enrichment Analysis
Functional enrichment analysis of DEmRNAs within the lncRNA-miRNA-mRNA triple regulatory network was conducted using the R packages “GOplot” and “clusterprofiler”. Based on the STRING database,17 a set of 61 PRRC2B-binding proteins, supported by experimental evidence, was obtained. Protein-protein interaction networks were visualized using the R package “igraph”, and GO enrichment (molecular function; MF, cellular component; CC, biological process; BP) and KEGG pathway analyses were performed and visualized using the R package “ggplot2”. A significance threshold of p < 0.05 was applied to GO terms and KEGG pathways.
The Receiver Operating Characteristic (ROC) Curves of Hub Genes
To evaluate the diagnostic value of hub genes, ROC curve analysis was performed using the R package pROC. The true positive rate (TPR) of sensitivity represented the number of true positive samples detected divided by the number of all true positive samples. The false-positive rate (FRP) represented the number of true negative samples detected divided by the number of all true negative samples. The ordinate represented sensitivity, while the abscissa represented 1 - specificity. The area under the ROC curve (AUC) was used as index of accuracy of prediction. The AUC represented the accuracy of prediction, categorized as low (0.5–0.7), moderate (0.7–0.9), or high (>0.9) predictive power.
Statistical Analysis
All statistical analyses were performed using R software and relevant packages. Statistical significance was represented as follows: ns (not significant, p > 0.05), *p < 0.05, **p < 0.01, ***p < 0.001.
Results
Identification of Differentially Expressed Genes (DEGs)
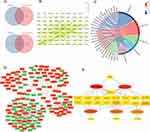
A total of 16 PBMCs from COPD patients (n = 8) and healthy controls (n = 8) were used for DElncRNA and DEmRNA screening, while 12 PBMCs from COPD patients (n = 6) and healthy controls (n = 6) were used for DEmiRNA identification. The criteria |log2FC| > 1.5 and p < 0.05 for lncRNAs (19 upregulated and 16 downregulated) and mRNAs (1145 upregulated and 528 downregulated), and |log2FC| > 0.5 and p < 0.05 for miRNAs (24 upregulated and 14 downregulated) were applied. Volcano plots and heatmaps were employed to visualize the distribution and expression of DEGs across PBMCs from COPD and healthy controls (Figure 1).
Construction of the lncRNA-miRNA-mRNA Triple Regulatory Network
DElncRNAs were subjected to the miRNet database to predict miRNAs targeting them, resulting in 255 predicted miRNAs. The intersection with DEmiRNAs yielded 5 selected miRNAs (Figure 2A). Then, these 5 miRNAs were then used to predict mRNA targets through the miRNet database, identifying 149 mRNAs through intersection with 1673 DEmRNAs (Figure 2A). A refined PPI network was created from the 149 identified genes, resulting in an 80-node, 104-edge network (Figure 2B). GO-CC enrichment analysis revealed enriched terms, including transcription regulatory complex (GO: 0005667), cell leading edge (GO: 0031252), endoplasmic reticulum-Golgi intermediate compartment (GO: 0005793), lamellipodium (GO: 0030027), and RNA polymerase II transcription regulator complex (GO: 0090575) (Figure 2C and Table 2). Finally, a lncRNA-miRNA-mRNA triple regulatory network encompassing 4 lncRNAs, 5 miRNAs, and 149 mRNAs was created using Cytoscape (Figure 2D).
 |
Table 2 GO-CC Enrichment Analysis of the Intersection Between DEmRNAs and Predicted mRNAs |
Hub triple regulatory network was determined, comprising 4 lncRNAs (MIAT, POLR2J4, C9orf163, and LINC00482), 5 miRNAs (hsa-miR-495-3p, hsa-miR-3127-5p, has-miR-199b-5p, hsa-miR-1287-5p, and hsa-miR-6088), and 17 mRNAs (PRRC2B, LENG8, PCGF5, PPM1F, JUNB, KLHL28, COMT, IRS2, STRN, MTA1, NAP1L1, ZNF512B, PATZ1, SKI, CMPK1, SPTLC2, and SLC30A9) (Figure 2E).
Construction of the ceRNA Network and Selection of a Model with a COPD-Specific Diagnostic Value
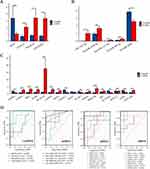
To establish a crucial ceRNA of great diagnostic value in COPD, expression levels of hub triple regulatory network RNAs in PBMCs from COPD and healthy controls were analyzed. Three upregulated (POLR2J4, C9orf163, and LINC00482) lncRNAs (Figure 3A), one downregulated (hsa-miR-6088) and two upregulated (hsa-miR-1287-5p and hsa-miR-3127-5p) miRNAs (Figure 3B), and eight upregulated (COMT, IRS2, JUNB, LENG8, MTA1, PPMIF, PRRC2B, and ZNF512B) and five downregulated (CMPK1, NAP1L1, PCGF5, SLC30A9, and STRN) mRNAs were identified (Figure 3C). Then, ROC curve analysis was performed, revealing diagnostic potential of 2 lncRNAs (C9orf163 and LINC00482), 2 miRNAs (hsa-miR-1287-5p and hsa-miR-6088), and 11 mRNAs (PCGF5, PRRC2B, SLC30A9, STRN, ZNF512B, MTA1, NAP1L1, COMT, IRS2, JUNB, and LENG8) with AUC above 0.9 (Figure 3D).
Given that cellular localization of lncRNAs determined the underlying mechanisms, we analyzed the subcellular localization of the three DElncRNAs by performing the lncLocator. As shown in Figure 4A, LINC00482 was mainly located in the cytoplasm, but the other two lncRNAs (POLR2J4 and C9orf163) was mainly distributed in the ribosome or exosome. These data indicate that LINC00482 may act as a ceRNA to modulate PRRC2B through sponging hsa-miR-6088. Thus, a LINC00482-miR-6088-PRRC2B ceRNA network was constructed (Figure 4B). In addition, the expression correlation analysis indicated a positive relationship between LINC00482 and PRRC2B expression in COPD (Figure 4C). Furthermore, the CTD database shows that LINC00482 and PRRC2B are associated with respiratory diseases (Figure 4D). Our validation with the GSE71220 dataset revealed higher PRRC2B expression in the whole blood of COPD patients compared to control subjects (Figure 4E).
PRRC2B-Related Functional Enrichment Analysis in COPD
To further screen out the targeted PRRC2B-binding proteins and investigate the molecular mechanism of PRRC2B in COPD, we acquired a total of 61 PRRC2B-binding proteins (supported by experimental evidence) from STRING database. Protein-protein interaction network visualization revealed interactions among these proteins (Figure 5A). GO enrichment analysis indicated involvement of PRRC2B-binding proteins in ribonucleoprotein granule, regulation of translation, and translation regulatory activity (Figure 5B–D). However, no significant KEGG pathway enrichment was observed in the PRRC2B-binding protein.
Discussion
COPD often goes undiagnosed and untreated until symptom worsen, and the pathological progression of the disease becomes evident. Therefore, there is an urgent need to explore rapid, cost-effective, and efficient clinical approaches for COPD. LncRNAs were once considered as “DNA junk” or “transcriptional noise”. However, recent research has unveiled their potential involvement in transcription, post-transcriptional processes, and intricate regulatory network, demonstrating multifaceted biological functions.18 Currently, the common regulatory mechanisms of lncRNAs on miRNAs have been validated. The mechanisms include acting as miRNA precursors,19 modulating transcription factor gene expression either as suppressors or activators,20 promoting primary miRNA processing,21 serving as “miRNA sponges” or “ceRNA” to competitively sequester miRNA response elements, among others. Among these mechanisms, the latter has emerged as a prominent research focus.22 In recent years, lncRNAs has been extensively reported to regulate the initiation progression of COPD by modulating various cellular processes, including proliferation, apoptosis, inflammation, migration, and epithelial-mesenchymal transition.23,24 In this study, we initially identified differentially expressed lncRNAs in PBMCs of COPD. Subsequently, we employed predicted algorithms to establish lncRNA-miRNA interactions. These predicted miRNAs were then intersected with differentially expressed miRNA within COPD patients’ PBMCs. The resulting intersected miRNAs were utilized to predict corresponding miRNA-mRNA pairs. Similarly, we conducted an intersection between the predicted mRNAs and differentially expressed mRNAs in COPD patients’ PBMCs. This multi-step process culminated in the construction of a comprehensive lncRNA-miRNA-mRNA triple regulatory network containing 4 lncRNAs, 5 miRNAs, and 149 mRNAs.
We constructed a PPI network of 149 mRNAs and found that ACTB (Beta-actin) and VEGFA (Vascular endothelial growth factor A) as the core genes interacting with most of the other genes. ACTB is an abundant and highly conserved cytoskeleton structural protein that widely distributed in all eukaryotic cells and expression level of ACTB is abnormally low in both bronchoalveolar lavage fluid cells and biopsy tissue of asthma patients.25 VEGFA exhibited abnormally low expression in COPD quadriceps biopsy specimens26 and involved in changes in the bronchial microvascular and airway inflammation in COPD progression.27 In contrast, we observed for the first time a substantial increase of ACTB and VEGFA in PBMCs of COPD. The conflicting results indicated that the regulation of ACTB and VEGFA gene expression may vary between different sample types. While the enrichment analyses using KEGG pathway, GO-BP, and GO-MF did not yield significant results for these 149 genes, the outcomes of the GO-CC enrichment analysis revealed that these genes are predominantly enriched cell movement and gene expression regulation. Given that the pathophysiology of COPD involves airway remodeling, inflammation, and impaired gene regulation,28 the GO-CC analysis underscores the potential role of these genes in contributing to the molecular mechanisms underpinning COPD progression. Next, a key triple regulatory network was obtained by hub analysis based on a score > 2, including 4 lncRNAs, 5 miRNAs, and 17 mRNAs. Finally, the expression analysis and ROC analysis were performed on this hub regulatory network. In addition, since the interaction in the ceRNA network is exclusively confined to the cytoplasm, we conducted subcellular analysis for the four types of lncRNAs within the network. Among COPD patients, LINC00482 expression was positively correlated with PRRC2B expression and CTD database also identified a relatively high score for LINC00482/PRRC2B in inflammatory. The elevated expression of PRRC2B in COPD patients was further corroborated by our analysis of the GSE71220 dataset. This supports the notion that PRRC2B plays a significant role in the pathophysiology of COPD. In summary, the LINC00482-has-miR-6088-PRRC2B overexpressed ceRNA network in PBMCs related to the diagnostic of COPD was obtained.
LINC00482, a new-found lncRNA that is highly expressed in many types of tumors. Liao et al29 found that LINC00482 acts as a ceRNA by binding to miR-2467-3p, thereby activating the Wnt/β-catenin signaling and promoting cancer bone metastasis. The findings of Wang et al30 revealed that LINC00482 interacts with FOXA1 to induce the expression of MMP15, consequently promoting inflammation and angiogenesis in bladder cancer, and Xu et al31 elucidated that LINC00482 competes for binding with miR-142-3p, resulting in the upregulation of miR-142-3p target gene TGFβ-1 expression in microglial cell line HMC3. This, in return, facilitates microglial M2 polarization and promotes non-small cell lung cancer brain metastasis.These studies indicate the involvement of LINC00482 in various cellular biological processes of tumor cells, yet its presence in non-tumor context remains unreported. On the other hand, hsa-miR-6088 has also received limited attention with its abnormal downregulation currently only associated with gliomas and gastrointestinal cancer development.32,33 It is noteworthy that abnormal downregulation of hsa-miR-6088 has also been detected in peripheral blood of systemic lupus erythematosus patients.34 In congruence with these findings, our results present a pioneering observation: the aberrant upregulation of LINC00482 in PBMCs of COPD patients, along with the abnormal downregulation of hsa-miR-6088. Additionally, we found LINC00482 functions as a ceRNA competitively binding hsa-miR-6088 and thereby regulating downstream PRRC2B expression, which offering a deeper understanding of the molecular underpinnings of COPD pathogenesis.
PRRC2B, has been identified as an mRNA binding protein based on its enrichment of oligonucleotide (dT) captured mRNA across cell types.35 Its hypothesized arginine-glycine-rich (RG) domain, demonstrated to interact with RNA, is enriched in this context.36 Previous studies suggest that PRRC2B is a component of the translation initiation complex mediated by eukaryotic initiation factor 4G2 (elF4G2).37 This proposition is founded on its coimmunoprecipitation with elFG2 and eukaryotic initiation factor 3 (eIF3) in mouse embryonic stem cells.38 However, the precise function of PRRC2B in translation regulation and its mRNA targets remains unclear. Recent findings indicate that PRRC2B is highly expressed in tumor cells.39,40 It is hypothesized that PRRC2B may facilitate a set of proteins associated with tumor proliferation by binding to sequences near translation sites rich in CU or GA.41 Consistent with these findings, elevated expression of PRRC2B was also identified in PBMCs from COPD patients. Furthermore, PPI analysis of PRRC2B binding partners revealed that elF3 subunits were the major proteins interacting with PRRC2B. Enrichment analysis also indicated a significant enrichment in protein translation regulation. Therefore, we speculate that LINC00482/hsa-miR-6088 might contribute to the upregulation of PRRC2B in PBMCs, ultimately leading to aberrant proliferation of PBMCs in COPD, triggering inflammation and an imbalance in immune response in COPD, although further investigations are needed.
Our study exhibits certain notable limitation. Firstly, the bioinformatics results were derived from PBMCs of a relatively small cohort of COPD patients. The limited sample size might restrict the generalizability of our findings. Additionally, due to the lack of comprehensive clinical information, we were unable to explore the correlation between hub genes and clinical characteristics. Another significant drawback is the absence of experimental validation for the expression and mechanistic roles of the hub genes. Subsequent in vivo and vitro validation experiments are essential to confirm their functions.
Conclusion
In summary, our study not only identified pivotal hub genes in PBMCs of COPD but also constructed a ceRNA regulatory network. This contributes to understanding the pathophysiological processes of COPD through bioinformatics analysis, expanding our knowledge of COPD, and providing a foundation for potential diagnostic and therapeutic targets for COPD.
Data Sharing Statement
The datasets generated and/or analyzed during the current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/).
Ethical Approval
This study was reviewed and approved by Medical Ethics Committee of Guangzhou Women and Children’s Medical Center Liuzhou Hospital. The study protocol was thoroughly reviewed by the committee, ensuring adherence to principles of safety and fairness. Throughout the research process, the right and well-being of participants were fully protected. The Ethics Committee granted permission for the conduct of this study.
Consent for Publication
All participating authors give their consent for this work to be published.
Acknowledgments
The authors would like to sincerely thank the GEO for data sharing. This work was supported by the Self-funded by the Health Commission of Guangxi Zhuang autonomous Region (Z20210585, Z20200704, and Z20200841).
Author Contributions
All authors have significantly contributed to this research, encompassing the conception, study design, execution, data acquisition, analysis, and interpretation. Each author was actively involved in drafting and critically reviewing the manuscript. We collectively approved the final version to be published, agreed upon the choice of journal for submission, and accept full accountability for all aspects of the work.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Chen S, Kuhn M, Prettner K, et al. The global economic burden of chronic obstructive pulmonary disease for 204 countries and territories in 2020–50: a health-augmented macroeconomic modelling study. Lancet Glob Health. 2023;11:e1183–e93. doi:10.1016/S2214-109X(23)00217-6
2. Christenson SA, Smith BM, Bafadhel M, Putcha N. Chronic obstructive pulmonary disease. Lancet. 2022;399:2227–2242. doi:10.1016/S0140-6736(22)00470-6
3. Suissa S, Dell’Aniello S, Ernst P. Long-term natural history of chronic obstructive pulmonary disease: severe exacerbations and mortality. Thorax. 2012;67:957–963. doi:10.1136/thoraxjnl-2011-201518
4. Hurst JR, Donaldson GC, Perera WR, et al. Use of plasma biomarkers at exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174:867–874. doi:10.1164/rccm.200604-506OC
5. Sivapalan P, Lapperre TS, Janner J, et al. Eosinophil-guided corticosteroid therapy in patients admitted to hospital with COPD exacerbation (CORTICO-COP): a multicentre, randomised, controlled, open-label, non-inferiority trial. Lancet Respir Med. 2019;7:699–709. doi:10.1016/S2213-2600(19)30176-6
6. Nuñez A, Marras V, Harlander M, et al. Association between routine blood biomarkers and clinical phenotypes and exacerbations in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2020;15:681–690. doi:10.2147/COPD.S240720
7. Aldonyte R, Jansson L, Piitulainen E, Janciauskiene S. Circulating monocytes from healthy individuals and COPD patients. Respir Res. 2003;4:11. doi:10.1186/1465-9921-4-11
8. Cornwell WD, Kim V, Fan X, et al. Activation and polarization of circulating monocytes in severe chronic obstructive pulmonary disease. BMC Pulm Med. 2018;18:101. doi:10.1186/s12890-018-0664-y
9. Bahr TM, Hughes GJ, Armstrong M, et al. Peripheral blood mononuclear cell gene expression in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2013;49:316–323. doi:10.1165/rcmb.2012-0230OC
10. Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;9:8. doi:10.3390/cells9010008
11. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi:10.1016/j.cell.2011.07.014
12. Chang L, Xia J. MicroRNA regulatory network analysis using miRNet 2.0. Methods Mol Biol. 2023;2594:185–204.
13. McGeary SE, Lin KS, Shi CY, et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;2019:366.
14. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–d31. doi:10.1093/nar/gkz757
15. Volders PJ, Anckaert J, Verheggen K, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47:D135–d9. doi:10.1093/nar/gky1031
16. Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34:2185–2194. doi:10.1093/bioinformatics/bty085
17. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–d46. doi:10.1093/nar/gkac1000
18. Chen L, Zhou Y, Li H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018;257:25–32. doi:10.1016/j.virusres.2018.08.018
19. Ulitsky I. Interactions between short and long noncoding RNAs. FEBS Lett. 2018;592:2874–2883. doi:10.1002/1873-3468.13085
20. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. doi:10.1126/sciadv.aao2110
21. Chen Z, Chen X, Lei T, et al. Integrative analysis of NSCLC identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. Mol Ther. 2020;28:1479–1493. doi:10.1016/j.ymthe.2020.03.010
22. Xu J, Xu J, Liu X, Jiang J. The role of lncRNA-mediated ceRNA regulatory networks in pancreatic cancer. Cell Death Discov. 2022;8:287. doi:10.1038/s41420-022-01061-x
23. Wang Y, Chen J, Chen W, et al. LINC00987 ameliorates COPD by regulating LPS-induced cell apoptosis, oxidative stress, inflammation and autophagy through let-7b-5p/SIRT1 axis. Int J Chron Obstruct Pulmon Dis. 2020;15:3213–3225. doi:10.2147/COPD.S276429
24. Qiao X, Hou G, He YL, et al. The novel regulatory role of the lncRNA-miRNA-mRNA axis in chronic inflammatory airway diseases. Front Mol Biosci. 2022;9:927549. doi:10.3389/fmolb.2022.927549
25. Glare EM, Divjak M, Bailey MJ, Walters EH. beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57:765–770. doi:10.1136/thorax.57.9.765
26. Yang D, Yan Y, Hu F, Wang T. CYP1B1, VEGFA, BCL2, and CDKN1A affect the development of chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2020;15:167–175. doi:10.2147/COPD.S220675
27. Gao X, Wang X, Jiao N, Chen J, Sun D. Association of VEGFA polymorphisms with chronic obstructive pulmonary disease in Chinese Han and Mongolian populations. Exp Physiol. 2021;106:1839–1848. doi:10.1113/EP089523
28. Hogg JC, Chu F, Utokaparch S, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–2653. doi:10.1056/NEJMoa032158
29. Liao S, Fang X, Zhou K, et al. LINC00482 sponged miR-2467-3p to promote bone metastasis of prostate cancer through activating Wnt/β-catenin signaling pathway. J Bone Oncol. 2023;41:100494. doi:10.1016/j.jbo.2023.100494
30. Wang Y, Zhang L, Wei N, Sun Y, Pan W, Chen Y. Silencing LINC00482 inhibits tumor-associated inflammation and angiogenesis through down-regulation of MMP-15 via FOXA1 in bladder cancer. Aging. 2021;13(2):2264–2278. doi:10.18632/aging.202247
31. Xu W, Patel N, Deng Y, Ding S, Wang T, Zhang H. Extracellular vesicle-derived LINC00482 induces microglial M2 polarization to facilitate brain metastasis of NSCLC. Cancer Lett. 2023;561:216146. doi:10.1016/j.canlet.2023.216146
32. Gong X and Huang M. Tumor-suppressive function of lncRNA-MEG3 in glioma cells by regulating miR-6088/SMARCB1 axis. Biomed Res Int. 2020;2020:1–15. doi:10.1155/2020/4309161
33. Chen J, Xia Y, Sui X, et al. Steviol, a natural product inhibits proliferation of the gastrointestinal cancer cells intensively. Oncotarget. 2018; 9(41):26299–26308. doi:10.18632/oncotarget.25233
34. Ishibe Y, Kusaoi M, Murayama G, et al. Changes in the expression of circulating microRnas in systemic lupus erythematosus patient blood plasma after passing through a plasma adsorption membrane. Ther Apher Dial. 2018;22(3):278–289. doi:10.1111/1744-9987.12695
35. Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi:10.1016/j.cell.2012.04.031
36. Chowdhury MN, The JH. RGG motif proteins: interactions, functions, and regulations. Wiley Interdiscip Rev RNA. 2023;14:e1748. doi:10.1002/wrna.1748
37. de la Parra C, Ernlund A, Alard A, Ruggles K, Ueberheide B, Schneider RJ. A widespread alternate form of cap-dependent mRNA translation initiation. Nat Commun. 2018;9:3068. doi:10.1038/s41467-018-05539-0
38. Sugiyama H, Takahashi K, Yamamoto T, et al. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2017;114:340–345. doi:10.1073/pnas.1617234114
39. He S, Yang L, Xiao Z, Tang K, Xu D. Identification of key carcinogenic genes in Wilms’ tumor. Genes Genet Syst. 2021;96:141–149. doi:10.1266/ggs.21-00015
40. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–w102. doi:10.1093/nar/gkx247
41. Jiang F, Hedaya OM, Khor E, Wu J, Auguste M, Yao P. RNA binding protein PRRC2B mediates translation of specific mRNAs and regulates cell cycle progression. Nucleic Acids Res. 2023;51:5831–5846. doi:10.1093/nar/gkad322
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.