Back to Journals » Infection and Drug Resistance » Volume 12
Identification and antifungal susceptibility profiles of Kodamaea ohmeri based on a seven-year multicenter surveillance study
Authors Zhou M , Yu S, Kudinha T , Xiao M, Wang H, Xu Y, Zhao H
Received 3 April 2019
Accepted for publication 20 May 2019
Published 12 June 2019 Volume 2019:12 Pages 1657—1664
DOI https://doi.org/10.2147/IDR.S211033
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Menglan Zhou,1–3 Shuying Yu,1–3 Timothy Kudinha,4 Meng Xiao,1,3 He Wang,1,3 Yingchun Xu,1,3 Hongmei Zhao5
1Department of Clinical Laboratory, Peking Union Medical College Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing 100730, People’s Republic of China; 2Graduate School, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing 100730, People’s Republic of China; 3Beijing Key Laboratory for Mechanisms Research and Precision Diagnosis of Invasive Fungal Diseases, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, 100730, People’s Republic of China; 4Central West Pathology Laboratory, Charles Sturt University, Orange, New South Wales, Australia; 5Department of Clinical Laboratory, The People’s Hospital of Liaoning Province, Liaoning 110016, People’s Republic of China
Background: Kodamaea ohmeri has been a rare fungal pathogen in the past decades but is now becoming more common in various invasive fungal diseases, with high mortality. There are limited data on the occurrence and distribution of K. ohmeri.
Methods: Sixty-two K. ohmeri isolates collected from 24 hospitals in China over a 7-year period were studied. Performance of three phenotypic methods in the identification of this organism was assessed against a gold standard, 26S rDNA sequencing. Original identification results submitted by the participating local hospitals were reviewed. The Sensititre YeastOne YO10 (SYY) was evaluated in determining the in vitro antifungal susceptibilities using standard broth microdilution method (BMD) as a reference, and essential agreement (EA) was calculated.
Results: Accurate species identification was achieved in 82.3% and 96.8% of the cases by Vitek 2 Compact and Vitek mass spectrometry (MS), respectively. For Bruker MS, 12.9% and 96.8% of the isolates were correctly identified to species level using the direct transfer and protein extraction methods, respectively. Only 29 (46.8%) isolates were initially correctly identified as K. ohmeri by the local hospitals. The highest misidentification rate (100%, 16/16) was observed in CHROMagar. According to BMD, the highest MIC90 was seen in fluconazole (8 μg/mL), followed by 1 μg/mL for micafungin, caspofungin, 5-fluorocytosine, and amphotericin B, 0.5 μg/mL for itraconazole, 0.25 μg/mL for posaconazole and voriconazole. Significant differences in EAs for different drugs were observed, ranging from 95.2% for amphotericin B to 22.6% for itraconazole between SYY and BMD.
Conclusion: Our study emphasizes the need for accurate identification of clinical K. ohmeri isolates and the importance of validating antifungal susceptibility by standard BMD.
Keywords: Kodamaea ohmeri, identification, antifungal susceptibility profiles, invasive fungal disease, surveillance
Introduction
Invasive fungal disease (IFD) is an important cause of morbidity and mortality in hospitalized patients.1 The China Hospital Invasive Fungal Surveillance Net (CHIF-NET) program was the first, and currently the largest, national surveillance program established to provide updated information on the epidemiology of invasive fungal infections in mainland China. It was initiated in 2009, and by the seventh surveillance year (2016), as many as 73 hospitals from 30 of the 34 provinces in China had participated, enabling collection of over 8,000 yeast isolates. Although Candida species remain the most common fungal pathogens worldwide, recent reports have highlighted the emergence of infections caused by less-common pathogenic yeasts.2,3 One such emerging pathogen is Kodamaea ohmeri.
K. ohmeri, previously known as Pichia ohmeri and Yamadazyma ohmeri, is an ascosporogenous yeast, and a teleomorph of Candida guilliermondii var. membranaefaciens, which has been commonly used in the food industry for the fermentation of pickles, rinds, and other fruit.4 Now, the genus Kodamaea is divided into 5 species (K. anthrophila, K. kakaduensis, K. laetipori, K. nitidulidarum, K. ohmeri) and only K. ohmeri shows pathogenicity in humans.4 Since the first case report of sepsis due to K. ohmeri,5 several case reports of various IFDs, including sepsis or fungemia,6,7 catheter-related bloodstream infection,8,9 peritonitis,10 and endocarditis,11,12 with high mortality due to K. ohmeri have gradually accumulated. Moreover, nosocomial outbreaks of K. ohmeri infection in the pediatric ward have also been reported.13 All this evidence suggests that K. ohmeri should be added to the growing list of opportunistic fungal pathogens in humans, and calls for early recognition and appropriate treatment.
Despite the increasing clinical significance in IFD, there are limited data on the occurrence and distribution of K. ohmeri globally. Here, we studied the epidemiology and antifungal susceptibility patterns of K. ohmeri clinical isolates based on the multicenter surveillance program-CHIF-NET in China over seven years.
Materials and methods
Ethi cs
The study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (no. S-263). Written informed consent was obtained from patients for the use of the samples in research.
Isolates
A total of 62 K. ohmeri clinical isolates collected from 24 different hospitals in 14 provinces, as part of the CHIF-NET study, from August 2009 to July 2016, were studied. The study inclusion criteria were as follows: for each surveillance year, all non-repetitive yeast isolates from eligible patients with IFDs were forwarded to the central laboratory, the Department of Clinical Laboratory, Peking Union Medical College Hospital (PUMCH), for species confirmative identification and antifungal susceptibility testing.
DNA extraction and identification
DNA extraction and amplification of the 26S ribosomal DNA were performed with primer pairs NL1/NL4, as previously described.14 The PCR products were sent to Riobiotech (Beijing, China) for sequencing. Identification was carried out by querying the sequences against GenBank database with nucleotide Basic Local Alignment Search Tool (BLASTn,
We also evaluated the performance of the Vitek-2 Compact (bioMérieux, France) and two MALDI-TOF MS systems, including the Vitek MS system (IVD Knowledgebase version 3.0; bioMérieux) and the Bruker Autoflex Speed TOF/TOF MS system (Biotyper version 3.1 software; Bruker Daltonics, USA) in the identification of K. ohmeri isolates. The Vitek-2 Compact Yeast card was used, and the final profile results were analyzed further as per the database specifications. For Vitek MS system, the results were scored in one of three ways as per the manufacturer’s recommendations.15 For Bruker MS, both direct transfer and the ethanol-formic acid protein extraction methods, as recommended by the manufacturer,16 were used for sample preparation. Identification was determined according to manufacturer-determined criteria: a score of <1.7 was interpreted as “no” identification, a score of 1.7–2.0 as identification to genus level, and a score of ≥2.0 as identification to species level.17
Antifungal susceptibility testing
In vitro susceptibilities of the isolates to eight antifungal drugs including amphotericin B, 5-flucytosine, fluconazole, itraconazole, voriconazole, posaconazole, micafungin, and caspofungin, were determined by broth microdilution method (BMD) as per Clinical and Laboratory Standards Institute (CLSI) guidelines (document M27-A3).18 Furthermore, we evaluated the performance of Sensititre YeastOne YO10 (SYY) (Thermo Scientific, USA) in antifungal susceptibility testing of K. ohmeri isolates as per the manufacturer’s instructions. For both methods, minimum inhibition concentrations (MIC) were read after 24 hrs incubation. Since there is neither clinical breakpoint (CBP) nor epidemiological cut-off values (ECVs) available for K. ohmeri, only essential agreement (EA) [percentage of MICs detected by SYY within a single doubling dilution of the corresponding BMD result] for each drug19 was calculated compared to BMD results (EA for anidulafungin was not calculated due to its inaccessibility in China). Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used as the quality control strains for identification and antifungal susceptibility testing.
Results
Detailed information of the study isolates is summarized in Table 1. The 62 isolates were collected from 62 patients at 24 hospitals located in 14 provinces across China over seven years. Thirty-four (34/62, 54.8%) of the strains were isolated in CHIF-NET year 2016, ten of which were isolated from one single hospital (Figure 1). The majority of the isolates were from patients admitted to surgical department (38.7%), medical department (32.3%), and intensive care unit (ICU) (14.5%), followed by pediatrics (6.5%), emergency (3.2%), organ transplantation (1.6%), dermatology (1.6%), and rehabilitation (1.6%). Among various specimen types, more than half of the isolates (54.8%) were recovered from blood, followed by catheter (16.1%), wound (8.1%), ascitic fluid (8.1%), drainage fluid (6.5%), broncho-alveolar lavage fluid (3.2%), pleural effusion (1.6%), and cerebrospinal fluid (1.6%) (Table 1).
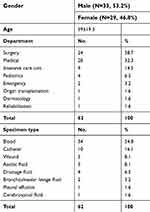 | Table 1 Distribution of the 62 Kodamaea ohmeri isolates by department and specimen type |
 | Figure 1 Distribution of 62 Kodamaea ohmeri isolated from the seven-year surveillance study.Abbreviation: CHIF-NET, China Hospital Invasive Fungal Surveillance Net. |
Sequence-based identification
The 26S rDNA sequences of the study isolates exhibited 99–100% identity with the sequence of standard strain CBS 6722 in the GenBank. The DNA sequences of the representative isolates have been deposited in GenBank with accession numbers MK414609 to MK414670.
Original species identification results by local hospitals
We looked back into the original information submitted by the local hospitals about the isolates and found that among the 62 K. ohmeri isolates re-identified by DNA sequencing in the central laboratory, only 29 (46.8%) were initially identified as K. ohmeri correctly by the local hospitals. The remaining 33 (53.2%) were misidentified as Candida albicans (n=11), Candida glabrata (n=9), Candida tropicalis (n=4), Candida guilliermondii (n=3), Candida lusitaniae (n=1), Cryptococcus neoformans (n=1), Candida famata (n=2), Candida pelliculosa (n=1) and Candida rugosa (n=1) by different methods. Vitek MS, Vitek 2 Compact, ATB32 C, and APC 20C correctly identified 100% (2/2), 70% (14/20), 66.7% (4/6), and 56.3% (9/16) of the isolates, respectively. Only one isolate each was identified using BD Phoenix100 and RapID™ YEAST PLUS, and neither of them got the correct result; one misidentified as C. tropicalis and the other one as Cryptococcus neoformans. Noticeably, the highest misidentification rate (16/16, 100%) was seen in CHROMagar among which nine, four, and three isolates were misidentified as Candida albicans, Candida glabrata, and Candida tropicalis, respectively (Table 2).
 | Table 2 Original identification results of the 62 Kodamaea ohmeri isolates submitted by local hospitals using different phenotypic methods |
Vitek 2 compact and MALDI-TOF MS identification results in the central laboratory
As compared with 26S rDNA sequencing, 82.3% of the isolates were correctly identified by Vitek 2 Compact system while 17.7% of the isolates yielded “no identification” results. Vitek MS system correctly identified 96.8% of the isolates (confidence value, 99.9%) with only one exception of “no identification” result. For the Bruker system, considerable differences were observed in the results of the two sample preparation methods used. According to the manufacturer-determined criteria, 12.9% and 71.0% of the isolates were identified to species and genus level, respectively, while 16.1% yielded “no identification” results using the direct transfer method. A significant increase in identification accuracy was seen when using the protein extraction method with 96.8% and 3.2% of the isolates correctly identified to species and genus levels, respectively (Table 3).
 | Table 3 Performance of Vitek 2 compact system, Vitek MS, and Bruker Biotyper MS compared with 26S rDNA gene sequencing for the identification of 62 Kodamaea ohmeri isolates |
Antifungal susceptibility profiles
The antifungal susceptibilities of the study isolates by BMD and SYY are shown in Table 4. Significant differences in EAs for different drugs were observed, ranging from 22.6% for itraconazole to 95.2% for amphotericin B. MIC50s and MIC90s detected by BMD were generally higher (up to four-fold) than those detected by SYY. Due to such a great inconsistency between BMD and SYY, which has been reported with high agreement in yeasts, we repeated the antifungal susceptibility testing by both methods to exclude experimental errors, and the previous results were confirmed. According to BMD, highest MIC90 was seen in fluconazole (8 μg/mL), followed by 1 μg/mL for micafungin, caspofungin, 5-fluorocytosine, and amphotericin B, 0.5 μg/mL for itraconazole, and 0.25 μg/mL for posaconazole and voriconazole.
 | Table 4 Comparison of in-vitro antifungal susceptibility data (MIC, μg/mL) of the 62 Kodamaea ohmeri isolates against nine antifungal agents between BMD and SYY |
Discussion
For decades, K. ohmeri has been recognized as a fungal contaminant but not as a human pathogen. Systemic infections due to K. ohmeri have generally been considered to be rare. Consequently, little attention has been paid to this insignificant yeast until 1998 when the first case of fungemia caused by K. ohmeri was described.5 Most published studies to date on K. ohmeri infections are sporadic cases commonly seen in Asian countries like Korea,20,21 Japan,8,22 and India.23–26 The infections were reported more often in children than in adults, and almost all patients had one or more underlying conditions alongside immunodeficiency.8 Mortality rates due to K. ohmeri invasive infections have been reported to be as high as 50%.13,20 The largest cluster of K. ohmeri infection reported was set in a single hospital in North India, presenting as 38 fungemia cases, 78.9% of which were isolated from neonates in intensive care units.26 Several surveillance studies in Spain, Malaysia, and Tunisia have also reported the isolation of K. ohmeri but with limited numbers.27–29 So far, our study presents as the first, largest, and multicenter epidemiological study of K. ohmeri clinical isolates causing IFDs in China.
During the 7-year surveillance, a total of 62 K. ohmeri isolates from cases of IFDs were collected and re-identified by 26S rDNA sequencing at the Central hospital, PUMCH. However, according to the original results submitted by the participating local hospitals, less than half of the isolates were correctly identified as K. ohmeri using different methods. The CHROMagar Candida chromogenic growth medium, which was developed based on the characteristic color change of the colonies, is an extremely useful tool in the clinical lab to assist in routine identification of common Candida species. However, this medium failed to correctly identify all the 16 K. ohmeri isolates reported by local hospitals in this study. It is known that K. ohmeri colonies can undergo a unique color change from pink to blue when grown on CHROMagar medium, and this phenomenon takes at least 2–3 days to form pink-blue colonies. Moreover, a full week may be required to obtain complete blue color colony development.20 Thus it is not surprising to note that 9, 4, and 3 isolates amongst the 16 misidentified isolates were incorrectly identified as C. albicans, C. glabrata, and C. tropicalis, respectively, which may be a result of determining the color of the isolates either too early or too late by lab staff. This identification confusion has been previously reported.26 Actually, this is how the largest cluster of K. ohmeri infection was discovered, in which 38 (25.7%) of 148 previously identified C. tropicalis isolates were re-identified as K. ohmeri by genotypic characterisation.26 Therefore, while CHROMagar is a useful and simple identification tool, careful and patient observation is necessary for correct identification of K. ohmeri.
Several other fungal identification methods with various levels of accuracy for specific organisms are used by many local hospitals in China. Among 16 isolates identified by API 20C system, seven (43.8%) were misidentified as C. glabrata (n=4), C. lusitaniae (n=1), C. albicans (n=1), and C. guilliermondii (n=1). The Vitek 2 Compact system correctly identified 70% (14/20) of isolates, with two misidentified as C. guilliermondii, two as C. famata, and one of each as C. pelliculosa and C. rugosa. Both the API 20C and Vitek 2 Compact systems have been shown to yield false-positive results, identifying C. haemulonii or C. parapsilosis as K. ohmeri.20,30 It is also known that C. auris and C. haemulonii are closely related and cannot be distinguished with conventional identification methods.31 Due to the emerging role of C. auris as a multidrug-resistant fungal pathogen with high morbidity and mortality, early accurate identification of this organism is crucial for patient management.32 In this case, the use of the faster molecular diagnostic tools for the proper identification of fungal pathogens is strongly recommended. The other four methods including ATB32C, BD Phoenix100, RapID™ YEAST PLUS, and Vitek MS, which were used for fungal identification by a small number of hospitals, yielded a wide variety of results which were difficult to generalize.
Nevertheless, the Vitek MS system performed the best among all the methods with 100% accuracy, albeit only two isolates identified using this method. Therefore, we performed a parallel study for identification of all the 62 K. ohmeri isolates using three commonly used phenotypic methods in our lab. To the best of our knowledge, this is the first evaluation study on the performance of three phenotypic methods for the identification of K. ohmeri. In general, the Vitek 2 Compact system (82.3% vs 70%) and Vitek MS (98.4% vs 100%) showed similar results with those submitted by the local hospitals. The Bruker system demonstrated a difference in the identification accuracy of K. ohmeri based on the sample preparation method. The identification accuracy obtained using the protein extraction method (96.8%) was comparable to that of Vitek MS, while the direct transfer method was only comparable to that of Vitek 2 Compact only when the genus identification cut-off value (>1.7) was adopted (83.9%). While MALDI-TOF MS is increasingly being used to identify Candida species in clinical laboratories, only one study has previously reported on the use of Bruker system for identification of K. ohmeri.8 In that study, the protein extraction procedure was used, which yielded a high confidence score although only one isolate was tested.8 These data suggest that both Vitek MS and Bruker system with protein extraction method for sample preparation can be used as a fast and accurate tool for K. ohmeri identification.
For the first time, we also compared the in-vitro susceptibilities of 8 antifungal agents using standard BMD and the commercial SYY against 62 K. ohmeri isolates. The commercial SYY system is an adapted microbroth susceptibility testing system based on the M27-A3 standard for yeasts, which has been widely used in antifungal susceptibility testing of yeasts and is now being evaluated in molds.33,34 Excellent EAs have been reported in triazoles and echinocandins against Candida spp., ranging from 92.3% to 100%.35,36 Surprisingly, significant differences in EAs against K. ohmeri isolates were observed, especially for itraconazole presenting as 22.6%. We excluded experimental errors by repeating both procedures, and the results were confirmed. We tried to find a possible explanation for this finding, but it proved difficult as there is limited data reported in literature on this aspect. Most of the published literature concerning K. ohmeri infections only used one method, either the commercial SYY or the standard BMD, to determine the MIC values. Only one previous study used BMD to confirm the MIC for micafungin using a colorimetric method for a single K. ohmeri isolate causing fungemia and found a 640-fold difference in the MIC value between the two methods.22 Despite the scarce evidence, this should serve as a reminder that validation of the antifungal susceptibility test by standard BMD for rare yeasts like K. ohmeri is important as susceptibility results often play an important role in the choice of antifungal agent to administer.
Since no CBP or ECV has been established to date for K. ohmeri, we could not compare the susceptibility or resistance patterns of our isolates with those reported in literature. However, based on MIC distribution of susceptibility results by BMD, our findings are in agreement with previous results showing that K. ohmeri strains have low MICs to all antifungal agents tested except for fluconazole, with MIC50 and MIC90 as high as 4 and 8 μg/mL, respectively.26 Twenty-four of the tested isolates exhibited MICs of ≥8 μg/mL, one of which had the highest MIC of >256 μg/mL. Compared to the large cluster from India,26 the present isolates had similar MIC90s for amphotericin B, itraconazole and posaconazole, while a two-fold increase in MIC90 was observed for voriconazole and caspofungin, and a four-fold increase for fluconazole. Previous studies concluded that amphotericin B or echinocandin should be considered to be a good antifungal choice for treatment of K. ohmeri infections.8,37 Considering that antifungal treatment should be adjusted according to susceptibility results of the clinical isolates, based on the MIC results of our isolates, all the eight antifungal agents tested except fluconazole, may be successful in treating most of the K. ohmeri infections according to the in vitro susceptibility results.
In conclusion, this is the first systemic study regarding the epidemiology, identification, and antifungal susceptibility profiles of K. ohmeri isolates in China. Our study emphasizes the need for accurate identification of clinical K. ohmeri isolates as an emerging human pathogen in China and the importance of validation of antifungal susceptibility results by standard BMD.
Acknowledgments
We thank all the laboratories that participated in the CHIF-NET program in 2010–2016. This work was supported by Graduate Innovation Fund of Peking Union Medical College (grant no. 2017-1002-1-21), Hebei Science and Technology Project (17277775D), and CAMS Initiative for Innovative Medicine (grant no. 2016-I2M-3-014). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
All authors report no conflicts of interest in this work.
References
1. Kullberg BJ, Arendrup MC. Invasive candidiasis. N Engl J Med. 2015;373(15):1445–1456. doi:10.1056/NEJMra1315399
2. Miceli MH, Diaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11(2):142–151. doi:10.1016/S1473-3099(10)70218-8
3. Pande A, Non LR, Romee R, Santos CA. Pseudozyma and other non-candida opportunistic yeast bloodstream infections in a large stem cell transplant center. Transpl Infect Dis. 2017;19(2):e12664. doi:10.1111/tid.12632
4. Kurtzman CP, Fell JW, Boekhout T. The Yeasts: A Taxonomic Study.
5. Bergman MM, Gagnon D, Doern GV. Pichia ohmeri fungemia. Diagn Microbiol Infect Dis. 1998;30(3):229–231.
6. Yang BH, Peng MY, Hou SJ, Sun JR, Lee SY, Lu JJ. Fluconazole-resistant Kodamaea ohmeri fungemia associated with cellulitis: case report and review of the literature. Int J Infect Dis. 2009;13(6):e493–e497. doi:10.1016/j.ijid.2009.02.003
7. Lee WG, Shin JH, Uh Y, et al. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 2011;49(9):3139–3142. doi:10.1128/JCM.00319-11
8. Kanno Y, Wakabayashi Y, Ikeda M, et al. Catheter-related bloodstream infection caused by Kodamaea ohmeri: a case report and literature review. J Infect Chemother. 2017;23(6):410–414. doi:10.1016/j.jiac.2017.01.003
9. Hitomi S, Kumao T, Onizawa K, Miyajima Y, Wakatsuki T. A case of central-venous-catheter-associated infection caused by Pichia ohmeri. J Hosp Infect. 2002;51(1):75–77. doi:10.1053/jhin.2002.1209
10. Choy BY, Wong SS, Chan TM, Lai KN. Pichia ohmeri peritonitis in a patient on CAPD: response to treatment with amphotericin. Perit Dial Int. 2000;20(1):91.
11. Joao I, Duarte J, Cotrim C, et al. Native valve endocarditis due to Pichia ohmeri. Heart Vessels. 2002;16(6):260–263. doi:10.1007/s003800200034
12. Reina JP, Larone DH, Sabetta JR, Krieger KK, Hartman BJ. Pichia ohmeri prosthetic valve endocarditis and review of the literature. Scand J Infect Dis. 2002;34(2):140–141.
13. Otag F, Kuyucu N, Erturan Z, Sen S, Emekdas G, Sugita T. An outbreak of Pichia ohmeri infection in the paediatric intensive care unit: case reports and review of the literature. Mycoses. 2005;48(4):265–269. doi:10.1111/j.1439-0507.2005.01126.x
14. Linton CJ, Borman AM, Cheung G, et al. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United kingdom mycology reference laboratory. J Clin Microbiol. 2007;45(4):1152–1158. doi:10.1128/JCM.02061-06
15. Zhang L, Xiao M, Wang H, et al. Yeast identification algorithm based on use of the Vitek MS system selectively supplemented with ribosomal DNA sequencing: proposal of a reference assay for invasive fungal surveillance programs in China. J Clin Microbiol. 2014;52(2):572–577. doi:10.1128/JCM.02543-13
16. Ghosh AK, Paul S, Sood P, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect. 2015;21(4):372–378. doi:10.1016/j.cmi.2014.11.009
17. Deak E, Charlton CL, Bobenchik AM, et al. Comparison of the Vitek MS and Bruker Microflex LT MALDI-TOF MS platforms for routine identification of commonly isolated bacteria and yeast in the clinical microbiology laboratory. Diagn Microbiol Infect Dis. 2015;81(1):27–33. doi:10.1016/j.diagmicrobio.2014.09.018
18.
19. Zhou M, Wang Y, Liu C, et al. Comparison of five commonly used automated susceptibility testing methods for accuracy in the China Antimicrobial Resistance Surveillance System (CARSS) hospitals. Infect Drug Resist. 2018;11:1347–1358. doi:10.2147/IDR.S166790
20. Lee JS, Shin JH, Kim MN, et al. Kodamaea ohmeri isolates from patients in a university hospital: identification, antifungal susceptibility, and pulsed-field gel electrophoresis analysis. J Clin Microbiol. 2007;45(3):1005–1010. doi:10.1128/JCM.02264-06
21. Shin DH, Park JH, Shin JH, Suh SP, Ryang DW, Kim SJ. Pichia ohmeri fungemia associated with phlebitis: successful treatment with amphotericin B. J Infect Chemother. 2003;9(1):88–89. doi:10.1007/s10156-002-0208-z
22. Tashiro A, Nei T, Sugimoto R, et al. Kodamaea ohmeri fungemia in severe burn: case study and literature review. Med Mycol Case Rep. 2018;22:21–23. doi:10.1016/j.mmcr.2018.07.005
23. Das K, Bhattacharyya A, Chandy M, et al. Infection control challenges of infrequent and rare fungal pathogens: lessons from disseminated Fusarium and Kodamaea ohmeri infections. Infect Control Hosp Epidemiol. 2015;36(7):866–868. doi:10.1017/ice.2015.103
24. Sundaram PS, Bijulal S, Tharakan JA, Antony M. Kodamaea ohmeri tricuspid valve endocarditis with right ventricular inflow obstruction in a neonate with structurally normal heart. Ann Pediatr Cardiol. 2011;4(1):77–80. doi:10.4103/0974-2069.79632
25. Menon T, Herrera M, Periasamy S, Palanivelu V, Sikhamani R, Wickes B. Oral candidiasis caused by Kodamaea ohmeri in a HIV patient in Chennai, India. Mycoses. 2010;53(5):458–459. doi:10.1111/j.1439-0507.2009.01731.x
26. Chakrabarti A, Rudramurthy SM, Kale P, et al. Epidemiological study of a large cluster of fungaemia cases due to Kodamaea ohmeri in an Indian tertiary care centre. Clin Microbiol Infect. 2014;20(2):O83–O89. doi:10.1111/1469-0691.12337
27. Fernandez-Ruiz M, Guinea J, Puig-Asensio M, et al. Fungemia due to rare opportunistic yeasts: data from a population-based surveillance in Spain. Med Mycol. 2017;55(2):125–136. doi:10.1093/mmy/myw055
28. Eddouzi J, Lohberger A, Vogne C, Manai M, Sanglard D. Identification and antifungal susceptibility of a large collection of yeast strains isolated in Tunisian hospitals. Med Mycol. 2013;51(7):737–746. doi:10.3109/13693786.2013.800239
29. Ng KP, Kuan CS, Kaur H, Na SL, Atiya N, Velayuthan RD. Candida species epidemiology 2000–2013: a laboratory-based report. Trop Med Int Health. 2015;20(11):1447–1453. doi:10.1111/tmi.12577
30. Rodero L, Cuenca-Estrella M, Cordoba S, et al. Transient fungemia caused by an amphotericin B-resistant isolate of Candida haemulonii. J Clin Microbiol. 2002;40(6):2266–2269. doi:10.1128/jcm.40.6.2266-2269.2002
31. Kathuria S, Singh PK, Sharma C, et al. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI Broth microdilution, and Etest method. J Clin Microbiol. 2015;53(6):1823–1830. doi:10.1128/JCM.00367-15
32. Jeffery-Smith A, Taori SK, Schelenz S, et al. Candida auris: a Review of the Literature. Clin Microbiol Rev. 2017;31(1):e00029–17. doi:10.1128/CMR.00029-17
33. Siopi M, Pournaras S, Meletiadis J. Comparative evaluation of sensititre YeastOne and CLSI M38-A2 reference method for antifungal susceptibility testing of Aspergillus spp. against Echinocandins. J Clin Microbiol. 2017;55(6):1714–1719. doi:10.1128/JCM.00044-17
34. Wang HC, Hsieh MI, Choi PC, Wu CJ. Comparison of the sensititre YeastOne and CLSI M38-A2 microdilution methods in determining the activity of amphotericin B, itraconazole, voriconazole, and posaconazole against Aspergillus Species. J Clin Microbiol. 2018;56(10):e00780–18. doi:10.1128/JCM.00780-18
35. Pfaller MA, Chaturvedi V, Diekema DJ, et al. Clinical evaluation of the sensititre YeastOne colorimetric antifungal panel for antifungal susceptibility testing of the echinocandins anidulafungin, caspofungin, and micafungin. J Clin Microbiol. 2008;46(7):2155–2159. doi:10.1128/JCM.00493-08
36. Pfaller MA, Espinel-Ingroff A, Jones RN. Clinical evaluation of the sensititre YeastOne colorimetric antifungal plate for antifungal susceptibility testing of the new triazoles voriconazole, posaconazole, and ravuconazole. J Clin Microbiol. 2004;42(10):4577–4580. doi:10.1128/JCM.42.10.4577-4580.2004
37. Distasi MA, Del Gaudio T, Pellegrino G, Pirronti A, Passera M, Farina C. Fungemia due to Kodamaea ohmeri: first isolating in Italy. Case report and review of literature. J Mycol Med. 2015;25(4):310–316. doi:10.1016/j.mycmed.2015.08.002
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
