Back to Journals » Journal of Inflammation Research » Volume 16
Identification and Analysis of Neutrophil Extracellular Trap-Related Genes in Osteoarthritis by Bioinformatics and Experimental Verification
Authors Luan T, Yang X , Kuang G, Wang T, He J , Liu Z, Gong X, Wan J, Li K
Received 19 April 2023
Accepted for publication 11 August 2023
Published 31 August 2023 Volume 2023:16 Pages 3837—3852
DOI https://doi.org/10.2147/JIR.S414452
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Ning Quan
Tiankuo Luan,1,* Xian Yang,2,* Ge Kuang,1 Ting Wang,3 Jiaming He,1 Zhibo Liu,3 Xia Gong,1 Jingyuan Wan,2 Ke Li4
1Department of Anatomy, Chongqing Medical University, Chongqing, People’s Republic of China; 2Department of Pharmacology, Chongqing Medical University, Chongqing, People’s Republic of China; 3Department of Orthopedics, the Second Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 4Department of Orthopedics, the First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jingyuan Wan; Ke Li, Email [email protected]; [email protected]
Background: Osteoarthritis (OA) is a common joint disease with long-term pain and dysfunction that negatively affects the quality of life of patients. Neutrophil extracellular traps (NETs), consisting of DNA, proteins and cytoplasm, are released by neutrophils and play an important role in a variety of diseases. However, the relationship between OA and NETs is unclear.
Methods: In our study, we used bioinformatics to explore the relationship between OA and NETs and the potential biological markers. GSE55235, GSE55457, GSE117999 and GSE98918 were downloaded from the Gene Expression Omnibus (GEO) database for subsequent analysis.After differential analysis of OA expression matrices, intersection with NET-related genes (NRGs) was taken to identify Differentially expressed NRGs (DE-NRGs) in OA processes. Evaluation of immune cell infiltration by ssGSEA and CIBERSORT algorithm. The GSVA method was used to analyze the activity changes of Neutrophils pathway, Neutrophil degranulation and Neutrophil granule constituents pathway.
Results: Based on RandomForest (RF), Least Absolute Shrinkage and Selection Operator (LASSO), and Support Vector Machine-Recursive Feature Elimination (SVM-RFE) learning algorithms, five core genes (CRISPLD2, IL1B, SLC25A37, MMP9, and TLR7) were identified to construct an OA-related nomogram model for predicting OA progression. ROC curve results for these genes validated the nomogram’s reliability. Correlation analysis, functional enrichment, and drug predictions were performed for the core genes. TLR7 emerged as a key focus due to its high importance ranking in RF and SVM-RFE analyses. Gene Set Enrichment Analysis (GSEA) revealed a strong association between TLR7 and the Neutrophil extracellular trap pathway. Expression of core genes was demonstrated in mice OA models and human OA samples. TLR7 expression in ATDC5 cell line was significantly higher than control after TNFα induction, along with increased IL6 and MMP13.
Conclusion: TLR7 may be related to NETs and affects OA.
Keywords: neutrophil extracellular traps, osteoarthritis, immune infiltration, TLR7, machine learning algorithms
Introduction
OA is a degenerative joint disease that primarily affects the elderly population. It is a multifactorial disorder with a complex pathogenesis, involving a variety of joint tissues. In addition to the well-established degradation of articular cartilage, OA encompasses a comprehensive joint pathology, encompassing the synovial membrane, subchondral bone, menisci, and infrapatellar fat pad. These joint components play pivotal roles in preserving joint homeostasis and function. Their alterations profoundly influence biomechanical changes, thus exerting a significant impact on the overall disease progression and its severity.1–4 The cartilage in the joints continues to suffer wear and tear as it functions as a cushion, which over time may lead to pain, stiffness, and limited range of motion.5 The following factors are strongly associated with osteoarthritis: age, obesity, genetic predisposition, and joint injury.6 In OA patients, the inflammatory response in the synovial membrane is driven by the release of damage-associated molecular patterns (DAMPs) during the degradation and breakdown of articular cartilage.7,8 The inflammatory response leads to cellular damage and tissue injury, further accelerating the destruction of joint structures. Older adults with OA experience difficulty with daily activities, decreased independence, and increased risk of falls and fractures. Chronic joint pain could also lead to a decrease in sleep quality and even anxiety, further reducing their physical function.9 However, there is a lack of clinically effective treatment options for OA, and the molecular mechanisms and triggering factors of the disease are not well understood.
Upon entry of bacteria or viruses into the body, neutrophils receive signals and activated. Activated neutrophils release Peptidyl arginine deiminase 4 (PAD4) into the nucleus, which induces the release of DNA and granulocyte material from the nucleus to the extracellular to form NETs.10 NETs have different roles in various diseases and are similar to a double-edged sword. The NETs were initially thought to be a special mechanism for protecting the host from pathogens and destroying them.11 However, NETs have recently been found to increase the inflammatory response in autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus.12 A significant increase in polymorphonuclear neutrophil [PMN]-NETs was observed in RA patients.13 Neutrophils in the synovial fluid of patients with RA are accompanied by high levels of expression of peptidylarginine deiminases during disease onset.14 Recent bioinformatics-based studies have found that genes associated with NETs can be of clinical value as prognostic for patients with pan-cancer and ischemia reperfusion injury, and have an intimate relationship with NETs.13,15 Yet, comprehensive and in-depth studies on OA and NETs are still lacking.
Our study attempts to comprehensively analyze the subtle relationships between OA, NRGs and NETs-related signaling pathways (Neutrophils pathway, Neutrophil degranulation and Neutrophil granule constituents pathway). Based on OA-related datasets in the GEO database and NRGs collected in the literature, DE-NRGs was obtained after analysis. The GSVA algorithm was used to observe changes in NETs-related signaling pathways in OA patients. The CIBERSORT algorithm to evaluate the percentage of immune cell infiltration in OA patients. Next, a machine learning algorithm was used to determine the strongly correlated DE-NRGs and a Nomogram was constructed to help the clinical diagnosis and treatment of OA. Finally, we examined the previous results through animal experiments (mouse OA model), human OA samples and in vitro cellular experiments. In this study, we systematically analyzed the relationship between OA and NETs to help clinicians to diagnose and treat OA based on NETs as an entry point.
Materials and Methods
Data Collection
GSE55235 (con=10, OA=10), GSE55457 (con=10, OA=10), GSE98918 (con=12, OA=12) and GSE117999 (con=10, OA=10) downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/) for subsequent analysis.To ensure the robustness of the analysis, the four datasets were combined and batch effects were removed before further investigation.NRGs (n=69) were collected through the relevant literature,15 with specific information in Supplementary Table S1.
Identification of DE-NRGs in Patients with OA
Combine GSE55235, GSE55457, GSE98918 and GSE117999 and remove the batch effect. The “limma” package was used to identify differentially expressed genes (DEGs) in OA patients, with Adjusted p-value< 0.05 and |logFC| > 0.585 as screening criteria.16 After intersecting DEGs with NRGs, we successfully obtained the DE-NRGs.
Construction of Nomogram Model Based on DE-NRGs
Construction of a Nomogram DE-NRGs using the “rms” R Package. We evaluate each gene based on the I nfluence of DE-NRGs on the outcome variable, and then add up the evaluations to obtain the overall score. Finally, the total score was followed to provide assistance in the clinical diagnosis of OA patients.
Receiver Operating Characteristics (ROC) Analysis of DE-NRGs
To verify the usefulness of the Nomogram constructed by DE-NRGs for clinical diagnosis, we used the “pROC” package to plot ROC curves to evaluate the accuracy and usefulness of the Nomogram model.17
CIBERSORT Assessment of Immune Cell Infiltration in OA
The CIBERSORT algorithm was applied to determine the difference in immune cell counts between the control and OA groups, and the results were represented using a violin plot.18
Gene Set Variation Analysis (GSVA)
GSVA algorithm can evaluate the biological pathways or genomic activity changes in the disease, which is important for the diagnosis and prognosis of the disease.19 We applied this technique to analyze changes in biological pathways associated with NETs in OA patients.
Identification of Core DE-NRGs
Five core DE-NRGs were filtered by the results of the LASSO, SVM-RFE and RF algorithms.20 The Lasso method involves a penalty applied to the absolute values of the coefficients in order to regulate their magnitude, leading to improved control against overfitting and heightened interpretability of the model.21 The SVM-RFE algorithm is used to identify the most important signature genes related to a disease by eliminating noisy or redundant features, reducing the dimensionality of the data and improving the accuracy of the model.22 The RF algorithm for regression is a machine learning technique that builds decision trees using random subsets of data and features. This reduces the correlation between trees and enhances the overall performance.
PPI Network (Protein-Protein Interaction) and Functional Enrichment Analysis
GeneMANIA (http://www.genemania.org) is a database that integrates information on genetic interactions, physical interactions, co-expression, co-localization, and gene enrichment analysis.23 GeneMANIA can be utilized for construct PPI network and to generate gene function predictions and identify genes with comparable effects. The analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was conducted using the R package “clusterprofiler”. The Gene Set Enrichment Analysis (GSEA) algorithm was used to explore the biological changes that were significantly different between OA patients (median TLR7 expression was used to divide OA patients into two groups).24
Collection of Human Tissue Samples
Human cartilage was obtained from patients undergoing total knee arthroplasty at the First Affiliated Hospital of Chongqing Medical University (Chongqing, China). Human OA cartilage was obtained from osteoarthritis patients undergoing total knee arthroplasty at the First Affiliated Hospital of Chongqing Medical University (Chongqing, China) (n=6). The medial part of the tibial plateau was used as the OA group, while the lateral tibial plateau with less cartilage destruction was used as the normal group (n=6). Collected human biological materials were approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (2023.4.03 No: K2023-087). Patients provided informed consent following the principles of the Declaration of Helsinki.
Experiment OA in Mice
Male C57BL/6J mice Aged 8–10 weeks (25–30g) were purchased from the Animal Center of Chongqing Medical University. Animals were provided with sufficient food and water, housed in a stable environment at 22–26 degrees Celsius, and maintained on a regular circadian rhythm. The animal experiments covered in the article were in strict accordance with the guidelines of the Chongqing Medical University Animal Care and Use Committee (2022 12.06 No: 2022–620). The animal experiments were approved by the Animal Care and Use Committee of Chongqing Medical University. We randomly divided the mice into two groups (n=5), the Sham group and the DMM group. Destabilization of the medial meniscus (DMM) surgery was used to induce OA in mice. The sham group was performed on the knee with no ligament transection. The mice were fed the same diet for 8 weeks after surgery until they were sacrificed and the joints were collected for histology.
Histological Assessment
Knee cartilage destruction was assessed by safranin-O staining and scored using the OARSI grading system.25 In brief, knee joints from mice and cartilage from human were fixed in 4% paraformaldehyde (PFA), decalcified in 14% EDTA, embedded in paraffin, and sectioned at 5-um thickness. The sections were deparaffinized in xylene, hydrated with graded ethanol, and finally stained with safranin O- fast green (Solarbio, Beijing, China).
Immunofluorescence
Joints from mice were fixed in 4% PFA for 14 h and decalcified in 14% EDTA for 3 d. Furthermore, the cartilage from human was also fixed in 4% PFA for 16 h and decalcified in 14% EDTA for 5 d. Afterward, the joints and cartilage were preserved in an optimum cutting temperature (OCT) compound, cut to a thickness of 10μm, and subjected to immunofluorescence using antibodies recognizing Toll-like receptor 7 (Boster, China, 1:100).The immunofluorescence protocol can be found in our previously published article.26
Cell Culture
The mouse chondrogenic cell line ATDC5 cells were seeded into Dulbecco's modified Eagle’s medium (DMEM, Gibco, USA) which was supplemented with 10% fetal bovine serum (Gibco, USA), penicillin (100 units/mL), and streptomycin (100 μg/mL). Following reaching confluency, the ATDC5 cells were pretreated for 24 hours with tumor necrosis factor ⍺ (TNF-⍺, Abcam, USA) (20 ng/mL) or phosphate buffer saline (PBS).
Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
The process of mRNA and cDNA was conducted following the previously described method.26 Primers of interest genes were designed using NCBI Primer-Blast software. Primer sequences in Supplementary Table S2.
Statistics
In this study, all expression matrix data were analysed using R software v4.1.2. The results of the experimental and control groups were analyzed by two-tailed Student’s t-test, and one-way ANOVA was performed for multiple comparisons (p < 0.05 = *p < 0.01 = **p < 0.001 = ***p < 0.0001 = ****p >0.05 = ns).
Results
DE-NRGs Identification in OA Patients
The four OA-related data collected in the GEO database were combined and batch effects were corrected to minimize errors in the subsequent analysis. P-adjust <0.05 and |logFC|>0.585 were used as filtering criteria for the difference analysis of Con and OA groups, and finally 133 down-regulated differentially expressed genes and 120 up-regulated differentially expressed genes (DEGs) were obtained (Figure 1A and Supplementary Table S3). As shown in the heatmap, the top 50 genes whose expression has been significantly up- or down-regulated are highlighted (Figure 1B). Seven DE-NRGs (TLR7, CRISPLD2, SLC25A37, IL1B, IL6, MMP9 and TLR8) implicated in the OA process were identified by intersecting 255 DEGs with 69 NRGs (Figure 1C). The GSVA algorithm analysis of the NETs lineage related pathways (Neutrophils pathway, Neutrophil degranulation and Neutrophil granule constituents.) showed that the pathway activity was significantly elevated in the OA group to the Con group (Figure 1D).
Differences in the Immune Characteristics of OA Patients
The CIBERSORT algorithm was used to explore the detail of immune cell infiltration abundance between the OA and Con groups. OA patients had significantly higher abundance of T-cells CD4 memory activated, Macrophages M0 and Mast cells resting than in the Con group. In contrast, Plasma cells, T cells CD4 memory resting, Monocytes, Mast cells activated had a higher percentage of infiltration in the Con group (Figure 2A and B).
Identification of Core DE-NRGs and Analysis of Interactions with Trait-Associated Genes
We integrated the results of three machine learning algorithms, LASSO, SVM- RFE, and RF, and determined the most critical DE-NRGs for the OA process. Based on the results of linear regression, LASSO establishes five important DE-NRGs (Figure 3A and B). The results of SVM- RFE and RF indicated that these five genes were ranked as the highest (Figure 3C–F and Supplementary Table S4). Using correlation analysis of these five DE-NRGs we found that TLR7 and MMP9 were significantly positively correlated, while SLC25A37 CRISPLD2 and IL1B were significantly negatively correlated (Figure 4A). Next, we constructed a PPI co-expression network associated with DE-NRG by the GeneMANIA tool and performed KEGG/GO enrichment analysis (Figure 4B–D). The KEGG results focused on MAPK signaling pathway and cytokine-cytokine receptor interactions. Biological pathways, molecular functional outcomes involving cytokine-mediated signaling pathways and binding of cytokine receptors. The results of cellular components suggest that collagen-containing extracellular matrix and secretory granular lumen may be involved.
NETs-Related Nomogram Mode Construction and Validation
We developed a NETs related Nomogram model related based on 5 DE-NRGs (TLR7, CRISPLD2, SLC25A37, IL1B, and MMP9) to provide physicians with a streamlined and reliable diagnostic tool (Figure 5A). The calibration curve analysis showed that the accuracy of the Nomogram model for DE-NRGs was very similar to the actual positivity rate (Figure 5B). The results of both the decision curve analysis and the clinical impact curve analysis demonstrated that our Nomogram model constructed based on DE-NRGs can offer a significant level of support for identifying OA, as evidenced in Figures 5C and D. The ROC curve in Figures 5E–I also displays that the classification performance of CRISPLD2, SLC25A37, IL1B, MMP9, and TLR7 is excellent, as indicated by the high Area Under the Curve (AUC) values. Finally, we also verified the expression of CRISPLD2, SLC25A37, IL1B, MMP9, and TLR7 in GSE55235 and showed that these 5 DE-NRGs were also significantly different (Figure S1).
Predicting DE-NRGs Targeted Drugs
We delved deeper into the potential drugs that can effectively target DE-NRGs through analysis of the DGIdb database and visualized the results with Cytoscape software (Figure 6). Regrettably, our analysis did not yield a prediction for a drug targeting CRISPLD2. We ultimately uncovered 34 drugs that could potentially impact DE-NRGs, among which were 4 drugs targeting TLR7, 1 drug targeting CRISPLD2, 28 drugs targeting IL1B, and 1 drug targeting MMP9. The findings suggest that HYDROXYCHLOROQUINE SULFATE can be used as an antagonist of TLR7, while IMIQUIMOD can be utilized as an agonist of TLR7. RILONACEPT and CANAKINUMAB can be utilized as inhibitors of IL1B. Specific information can be found in Supplementary Table S5.
 |
Figure 6 Prediction of DE-NRGs targeted drugs. Network diagram showing that the DGIdb database predicts DE-NRGs related drugs. |
ssGSEA and GSEA to Explore the Formation of NETs
To determine whether there are significant differences in the activities of Neutrophils pathway, Neutrophil degranulation and Neutrophil granule constituents pathway, we performed validation in the GSE55235 dataset using the ssGSEA algorithm. The activity of Neutrophils pathway, Neutrophil degranulation and Neutrophil granule constituents pathway in OA group was significantly higher than that in Con group (Figure 7A). Results indicated a significant positive correlation between TLR7 and the Neutrophil degranulation and Neutrophil granule constituents pathway. Additionally, upregulation of MMP9 may boost the activity of Neutrophils pathway, Neutrophil degranulation and Neutrophil granule constituents pathway. CRISPLD2 and IL1B showed a significant negative correlation with the Neutrophil degranulation and Neutrophil granule constituents pathway. SLC25A37 was only negatively correlated with the Neutrophil granule constituents pathway (Figure 7B). Subsequently, we performed gene enrichment analysis on TLR7 in the GSE55235 and GSE55457 datasets. The results showed that TLR7 was enriched in the Neutrophil extracellular trap formation pathway in both GSE55235 and GSE55457 datasets (Figure 7C and D).
Validation of Bioinformatics Results by qPCR and Immunofluorescence
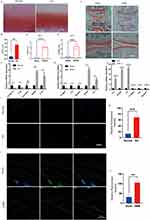
To elucidate the contribution of TLR7, CRISPLD2, SLC25A37, IL1B, and MMP9 in OA, we started experiments to verify the authenticity of human OA samples and established mouse OA models by staining with Safranin O-Fast Green (Figure 8A–C). This step was essential to ensure the reliability of the samples used in our investigation. Subsequently, the mouse OA model underwent medial tibial plateau (MTP) and medial femoral condyle (MFC) scoring to further reinforce the validity of the model (Figure 8D and E). The qPCR results showed that TLR7, CRISPLD2, IL1B, and MMP9 were significantly different in mRNA expression levels and were consistent with the bioinformatics analysis (Figure 8F and G). However, the difference in SLC25A37 mRNA expression was only observed in the mouse OA model (Figure 8F and G). As both the SVM-RFE and RF algorithms demonstrated that TLR7 was the most critical factor, we subsequently focused on analyzing TLR7. In the ATDC5 cell experiment, TNF-α stimulation led to a substantial increase in mRNA expression levels of TLR7, MMP9, and IL6, while CRISPLD2, IL1B, and SLC25A37 were notably downregulated (Figure 8H). Finally, we found that TLR7 expression was elevated in both human OA samples and mouse OA samples by immunofluorescence assay (Figure 8I–L).
Discussion
OA is a widespread degenerative joint condition that results in the deterioration of articular cartilage and bone. The progression of OA is intricately associated with aging, joint injury, and muscle wasting around the joints, alongside inflammation of the synovial membrane, degeneration of the infrapatellar fat pad, and meniscal degeneration.27 Approximately 60% of the elderly population is estimated to be affected by OA, causing significant degradation in their quality of life. The OA process has been suggested to be impacted by various cellular death mechanisms such as apoptosis, necroptosis, and cuproptosis,28 but the contribution of NETs to OA remains unclear.
NETs are composed of decondensed chromatin fibers and granule proteins such as histones and neutrophil elastase, which are released by activated neutrophils during degranulation.29 NETs can trap and kill pathogens, but they can also contribute to tissue damage in certain pathologies, such as autoimmune diseases and cancer. Neutrophils, which contain specialized organelles called neutrophil granules.30 These granules harbor a diverse range of molecules, including enzymes, antimicrobial peptides, and various proteins, enabling them to play a significant role in the body’s immune response. Upon encountering a pathogen, neutrophils activate the release of the granules they contain, thereby discharging their contents into the surrounding area and effectively combating the pathogen.
At the outset, we conducted a comprehensive analysis by merging four OA-related datasets, effectively correcting for batch effects, and subsequently identifying 253 DEGs with regulatory relevance, while comparing the control and OA groups. Our analysis resulted in the identification of 7 DE-NRGs including TLR7, SLC25A37, IL11B, CRISPLD2, MMP9, TLR8, and IL6. GSVA evaluates the effect of specific genomic alterations on disease by assigning scores based on the activity of gene sets involved in particular biological processes or functions.31 Using GSVA, we found that the gene sets for Neutrophils pathway, Neutrophil degranulation and Neutrophil granule constituents pathway were significantly more activated in OA group compared to the control group. Next, we used the CIBERSORT algorithm to analyze immune cell infiltration in OA patients. We discovered that plasma cells, monocytes, and activated mast cells were significantly more infiltrated in the control group compared to the OA group, while T cells CD4 memory resting, T cells CD4 memory activated, Macrophages M0, and resting mast cells were significantly more infiltrated in the OA group compared to the control group.
Machine learning algorithms offer various advantages, including improved model accuracy, reduced overfitting, enhanced interpretability, and effective handling of high-dimensional data.32 In the field of biomedical research, it is possible to more accurately predict core molecules in disease processes and identify potential clinical features.
The elevated expression of TLR7 in the blood of RA patients makes the synovial tissue more susceptible to reacting to citrullinated proteins and nucleic acids that are released into the extracellular environment through the formation of NETs.33 The research findings demonstrate a sustained down-regulation of SCL25A37 in individuals with Major Depressive Disorder (MDD), suggesting its potential utility as a biomarker for MDD diagnosis and a promising target for future therapeutic and diagnostic interventions.34 CRISPLD2 is a gene that is differentially expressed in both polycystic ovary syndrome and ovarian cancer and has significant clinical implications for the prognosis of ovarian cancer.35 The levels of serum MMP9 expression in OA patients increase significantly after total joint arthroplasty treatment, reaching a stable level by the fifth day. Our findings align with this observation, indicating the potential utility of detecting MMP9 expression in both blood and joint samples. Such assessments could offer valuable guidance to surgeons, enabling them to refine treatment strategies and mitigate surgical risks, ultimately leading to improved patient outcomes.36 Based on the expression of five DE-NGRs, we created NETs-related nomograms for the purpose of facilitating clinical risk assessment and treatment planning in OA patients. The follow-up five DE-NGR highly related genes were subjected to KEGG and GO analysis.
The KEGG results were most closely related to interactions between cytokine receptors and cytokines, osteoclast differentiation, and MAPK signaling. The biological processes, cellular components, and molecular functional analysis results are related to cytokine-mediated signaling pathways, the extracellular matrix containing collagen, and binding of cytokine receptors, respectively. During the progression of OA, cytokine-cytokine receptor interactions may lead to activation of the MAPK signaling pathway, which then promotes the differentiation of osteoclasts. This increased osteoclast activity can promote destruction of articular cartilage and bone, leading to the characteristic symptoms of OA, such as pain, stiffness, and decreased mobility.37 Additionally, the MAPK signaling pathway plays a role in regulating cytokine-mediated signaling pathways, including those that drive inflammation by producing cytokines.38 Because both SVM-RFE, and RF analyses showed that TLR7 was the top ranked DE-NRGs, we performed a single gene GSEA on TLR7. In GSE55235 and GSE554757, TLR7 was significantly enriched in the Neutrophil extracellular trap formation. Not only that, TLR7 was also significantly correlated with Neutrophil degranulation and Neutrophil granule constituents progression. Thus, we speculate that TLR7 may influence the OA process by activating the Neutrophil extracellular trap formation pathway and promoting the formation of NETs. We confirmed the effect of TLR7 on OA by constructing mouse OA models, human OA samples, and cellular experiments. The results consistently showed that TLR7 expression was significantly increased in OA.
However, our study does have certain limitations. Firstly, the relatively small sample size of the dataset may introduce some bias in our experimental results. To address this, future studies with larger sample sizes and advancements in sequencing technology will allow us to build more accurate prognostic models. Secondly, we acknowledge the need for further investigations into the specific mechanisms through which TLR7 influences the formation of NETs. In conclusion, our research offers valuable insights into the diagnosis and treatment of OA through targeted NETs, while recognizing the potential for future research to expand and deepen our understanding in this field.
Conclusions
In summary, our study comprehensively explored the association between the NETs process and OA and finally identified the hub genes. Next, the hub genes were combined with immune cells, NETs formation and gene-targeted drugs for an integrated analysis. Finally, TLR7 was found to potentially induce the formation of NETs to drive the progression of OA.
Data Sharing Statement
The article contains the data that was utilized to substantiate the conclusions of this research.
Institutional Review Board Statement
The human samples used in this study conformed to national and institutional ethical guidelines and were approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (2023.4.03 No: K2023-087). All animal experiments were in line with the Guide for the Animal Care and Use Committee of Chongqing Medical University.
Author Contributions
Each author significantly contributed to various aspects of the reported work, including conception, study design, data acquisition, analysis, interpretation, drafting, revising, and critical review of the article. All authors provided final approval for the submitted version and the selected journal, demonstrating their commitment to the entire research process and assuming accountability for its entirety.
Funding
This work was supported by the National Natural Science Foundation of China No.81902293(Ke. Li).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Battistelli M, Favero M, Burini D, et al. Morphological and ultrastructural analysis of normal, injured and osteoarthritic human knee menisci. Eur J Histochem. 2019;63:2998. doi:10.4081/ejh.2019.2998
2. Donell S. Subchondral bone remodelling in osteoarthritis. EFORT Open Rev. 2019;4:221–229. doi:10.1302/2058-5241.4.180102
3. Favero M, El-Hadi H, Belluzzi E, et al. Infrapatellar fat pad features in osteoarthritis: a histopathological and molecular study. Rheumatology. 2017;56:1784–1793. doi:10.1093/rheumatology/kex287
4. Poole AR. Osteoarthritis as a whole joint disease. HSS J. 2012;8:4–6. doi:10.1007/s11420-011-9248-6
5. Martel-Pelletier J, Barr AJ, Cicuttini FM, et al. Osteoarthritis. Nat Rev Dis Primers. 2016;2:16072. doi:10.1038/nrdp.2016.72
6. Wang T, He C. Pro-inflammatory cytokines: the link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 2018;44:38–50. doi:10.1016/j.cytogfr.2018.10.002
7. Barnett R. Osteoarthritis. Lancet. 2018;391:1985. doi:10.1016/S0140-6736(18)31064-X
8. Yao Q, Wu X, Tao C, et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal Transduct Target Ther. 2023;8:56. doi:10.1038/s41392-023-01330-w
9. Tore NG, Oskay D, Haznedaroglu S. The quality of physiotherapy and rehabilitation program and the effect of telerehabilitation on patients with knee osteoarthritis. Clin Rheumatol. 2023;42:903–915. doi:10.1007/s10067-022-06417-3
10. Liu X, Arfman T, Wichapong K, Reutelingsperger CPM, Voorberg J, Nicolaes GAF. PAD4 takes charge during neutrophil activation: impact of PAD4 mediated NET formation on immune-mediated disease. J Thromb Haemost. 2021;19:1607–1617. doi:10.1111/jth.15313
11. Song YH, Wang ZJ, Kang L, et al. PADs and NETs in digestive system: from physiology to pathology. Front Immunol. 2023;14:1077041. doi:10.3389/fimmu.2023.1077041
12. Lazar S, Kahlenberg JM. Systemic lupus erythematosus: new diagnostic and therapeutic approaches. Annu Rev Med. 2023;74:339–352. doi:10.1146/annurev-med-043021-032611
13. Liu X, Huo Y, Zhao J, et al. Endothelial cell protein C receptor regulates neutrophil extracellular trap-mediated rheumatoid arthritis disease progression. Int Immunopharmacol. 2022;112:109249. doi:10.1016/j.intimp.2022.109249
14. Spengler J, Lugonja B, Ytterberg AJ, et al. Release of active peptidyl arginine deiminases by neutrophils can explain production of extracellular citrullinated autoantigens in rheumatoid arthritis synovial fluid. Arth Rheumatol. 2015;67:3135–3145. doi:10.1002/art.39313
15. Zhang Y, Guo L, Dai Q, et al. A signature for pan-cancer prognosis based on neutrophil extracellular traps. J Immunother Cancer. 2022;10:e004210. doi:10.1136/jitc-2021-004210
16. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi:10.1093/nar/gkv007
17. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi:10.1186/1471-2105-12-77
18. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi:10.1038/nmeth.3337
19. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
20. Li Y, Qi D, Zhu B, Ye X. Analysis of m6A RNA methylation-related genes in liver hepatocellular carcinoma and their correlation with survival. Int J Mol Sci. 2021;22:1474. doi:10.3390/ijms22031474
21. Cao X, He J, Chen A, et al. Comprehensive analysis of necroptosis landscape in skin cutaneous melanoma for appealing its implications in prognosis estimation and microenvironment status. J Pers Med. 2023;13:245. doi:10.3390/jpm13020245
22. Xia J, Sun L, Xu S, et al. A model using support vector machines recursive feature elimination (SVM-RFE) Algorithm to Classify Whether COPD patients have been continuously managed according to GOLD guidelines. Int J Chron Obstruct Pulmon Dis. 2020;15:2779–2786. doi:10.2147/COPD.S271237
23. Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46:W60–w64. doi:10.1093/nar/gky311
24. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi:10.1073/pnas.0506580102
25. Tang J, Su N, Zhou S, et al. Fibroblast growth factor receptor 3 inhibits osteoarthritis progression in the knee joints of adult mice. Arthritis Rheumatol. 2016;68:2432–2443. doi:10.1002/art.39739
26. Hu J, Du H, Yuan Y, et al. MFG-E8 knockout aggravated nonalcoholic steatohepatitis by promoting the activation of TLR4/NF-κB signaling in mice. Mediators Inflamm. 2022;2022:1–13. doi:10.1155/2022/5188895
27. Sun X, Zhen X, Hu X, et al. Osteoarthritis in the middle-aged and elderly in china: prevalence and influencing factors. Int J Environ Res Public Health. 2019;16:4701. doi:10.3390/ijerph16234701
28. Yang J, Hu S, Bian Y, et al. Targeting cell death: pyroptosis, ferroptosis, apoptosis and necroptosis in osteoarthritis. Front Cell Dev Biol. 2021;9:789948. doi:10.3389/fcell.2021.789948
29. Fousert E, Toes R, Desai J. Neutrophil extracellular traps (NETs) take the central stage in driving autoimmune responses. Cells. 2020;9:915. doi:10.3390/cells9040915
30. Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–691. doi:10.1083/jcb.201006052
31. Ferreira MR, Santos GA, Biagi CA, Silva Junior WA, Zambuzzi WF. GSVA score reveals molecular signatures from transcriptomes for biomaterials comparison. J Biomed Mater Res A. 2021;109:1004–1014. doi:10.1002/jbm.a.37090
32. Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284:603–619. doi:10.1111/joim.12822
33. Abdelwahab A, Palosaari S, Abdelwahab SA, et al. Differential synovial tissue expression of TLRs in seropositive and seronegative rheumatoid arthritis: a preliminary report. Autoimmunity. 2021;54:23–34. doi:10.1080/08916934.2020.1864729
34. Huo YX, Huang L, Zhang DF, et al. Identification of SLC25A37 as a major depressive disorder risk gene. J Psychiatr Res. 2016;83:168–175. doi:10.1016/j.jpsychires.2016.09.011
35. Zou J, Li Y, Liao N, et al. Identification of key genes associated with polycystic ovary syndrome (PCOS) and ovarian cancer using an integrated bioinformatics analysis. J Ovarian Res. 2022;15:30. doi:10.1186/s13048-022-00962-w
36. Slovacek H, Khanna R, Poredos P, et al. Interrelationship of MMP-9, Proteoglycan-4, and inflammation in osteoarthritis patients undergoing total hip arthroplasty. Clin Appl Thromb Hemost. 2021;27:1076029621995569. doi:10.1177/1076029621995569
37. Malemud CJ. Negative Regulators of JAK/STAT signaling in rheumatoid arthritis and osteoarthritis. Int J Mol Sci. 2017;18:484. doi:10.3390/ijms18030484
38. Wei J, Gao C, Hu K, et al. Knockdown of DAPK1 attenuates IL-1β-induced extracellular matrix degradation and inflammatory response in osteoarthritis chondrocytes via regulating the p38 MAPK-signaling pathway. Allergol Immunopathol. 2022;50:169–175. doi:10.15586/aei.v50i6.744
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.