Back to Journals » Drug Design, Development and Therapy » Volume 16
Icariin and Competing Endogenous RNA Network: A Potential Protective Strategy Against Contrast-Induced Acute Kidney Injury
Authors Lin Y , Zhu G, Li X, Yu H, Luo Y, Lin J , Li R, Huang Z
Received 12 April 2022
Accepted for publication 10 July 2022
Published 23 July 2022 Volume 2022:16 Pages 2343—2363
DOI https://doi.org/10.2147/DDDT.S369100
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianbo Sun
Yan Lin,1,2,* Gaofeng Zhu,3,* Xiaoyong Li,4,* Huaxiao Yu,5 Yuhang Luo,5 Jiaqiong Lin,6 Renyuan Li,7 Zena Huang2
1Department of Nephrology, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China; 2Yunkang School of Medicine and Health, Nanfang College, Guangzhou, People’s Republic of China; 3Department of General Medicine, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, People’s Republic of China; 4Department of General Surgery, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China; 5The Third Clinical School, Guangzhou Medical University, Guangzhou, People’s Republic of China; 6Affiliated Dongguan Maternal and Child Healthcare Hospital, Southern Medical University, Guangzhou, People’s Republic of China; 7Department of Endocrinology and Metabolism, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zena Huang, Yunkang School of Medicine and Health, Nanfang College, Guangzhou, People’s Republic of China, Tel +86-13570466614, Email [email protected] Renyuan Li, Department of Endocrinology and Metabolism, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China, Tel +86-18926146852, Email [email protected]
Background: Icariin presents protective effect in several kidney diseases. However, the role of icariin in contrast-induced acute kidney injury (CIAKI) is still unclear. This study aimed to investigate the effect of icariin in CIAKI, as well as exploring the underlying mechanism from the aspect of interaction between protein-coding genes and non-coding RNAs.
Methods: The effect of icariin was evaluated in both in vivo and in vitro CIAKI models. Rat kidneys were collected for genome-wide sequencing. The differentially expressed genes (DEGs) were screened and visualized by R software. The function annotation of DEGs was analyzed by Metascape. By Cytoscape software, the competing endogenous RNA (ceRNA) network was constructed, and hub genes were selected. Expressions of hub genes were validated by PCR. Association of hub genes in the ceRNA network and renal function was also examined.
Results: Icariin protected against CIAKI in both in vivo and in vitro models. Based on DEGs in icariin pretreated CIAKI rats, lncRNA- and circRNA-associated ceRNA networks were constructed, respectively. Function annotation showed the ceRNA networks were enriched in ERK1 and ERK2 cascade, MAPK signaling and NF-κB signaling. Further, two circRNAs, six lncRNAs, four miRNAs and nine mRNAs were selected as hub genes of the ceRNA network. Among them, eight mRNAs (Acot1, Cbwd1, Ly6i, Map3k14, Mettl2b, Nyap1, Set and Utp20) were negatively correlated with renal function, while one mRNA (Tmem44) was positively correlated with renal function.
Conclusion: Icariin presented a protective effect against CIAKI. The ceRNA network, involving Acot1, Cbwd1, Ly6i, Map3k14, Mettl2, Nyap1, Set, Tmem44 and Utp20, might partially contribute to the underlying mechanism of icariin protection by regulation of ERK1 and ERK2 cascade, MAPK signaling and NF-κB signaling.
Keywords: icariin, contrast medium, acute kidney injury, competing endogenous RNA, non-coding RNA
Background
Contrast induced acute kidney injury (CIAKI) is characterized by development of acute kidney injury (AKI) after intravascular administration of contrast medium (CM).1 The Kidney Disease Improving Global Outcome (KDIGO) working group suggested a definition based on a plasma creatinine (Cr) level or a urinary volume, which is increase of Cr by no less than 1.5 times within 7 days or by at least 0.3mg per deciliter within 48 hours after exposure to CM, or decrease of urinary volume to less than 0.5 mL per kilogram of body weight per hour that persists for at least 6 hours after exposure.2 Many studies have shown that CIAKI is associated with severe adverse outcomes, including increased mortality3 and progression of underlying chronic kidney disease.4 However, preventive strategies of CIAKI in current practice are far from satisfying. The benefits of most pharmaceutical agents are still unconvincing,5 and the protective effect of intravenous fluids is also challenged by a recent study.6 Thus, it is urgent to develop a reliable preventive strategy for CIAKI in the large and growing population undergoing contrast enhanced procedures.
Epimedii Herba is a herbal medicine originating from Epimedium koreanum Nakai, Epimedium brevicornum Maximowicz, Epimedium pubescens Maximowicz, or Epimedium sagittatum Maximowicz. It is treated as a “kidney-tonifying” agent in traditional Chinese medicine.7 Icariin, a major flavonoid responsible for the pharmacological actions of Epimedii Herba,8 has shown promising protection in several kidney diseases, including cisplatin-induced cytotoxicity,9 ureteral obstruction10 and diabetic nephropathy.11 Icariin carries out renal protection by multiple biological activities, such as anti-inflammation,12 anti-oxidative stress,9,11 restoring autophagy11 and inhibition of NF-κB pathway.13 However, the potential effect of icariin in CIAKI has not been investigated so far.
Non-coding RNAs (ncRNAs) are transcripts with no or low coding potential, among which microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) attract most attention in current researches. Emerging evidence has indicated that ncRNAs are important in renal biological maintenance and pathophysiological process.14 LncRNAs, circRNAs and mRNAs may compete to bind to same miRNA, an association known as the competing endogenous RNA (ceRNA) mechanism.15 An amount of studies has shown that, by forming a ceRNA network with mRNAs, ncRNAs can affect the pathogenesis and therapeutic response of kidney diseases. Several ceRNA networks are identified in CIAKI rat models,16 while the underlying mechanism of ceRNA networks in prevention of CIAKI is still ambiguous.
In the present study, we first investigated the effect of icariin in CM treated rats and human renal tubular epithelial cells. Then we analyzed the expression profile and function annotation of mRNAs, miRNAs, lncRNAs and circRNAs based on next generation RNA sequencing. Further, we constructed an icariin-associated ceRNA network and identified key modules in the network. Finally, we assessed the correlation of key modules in ceRNA network and renal function. Our findings revealed that icariin showed profound protection against CIAKI via ceRNA network, providing a potential preventive strategy for CIAKI.
Methods
Materials
Icariin, with a purity of 98%, was purchased from Glpbio Technology (CA, USA, Cat. No.: GN10278). A type of nonionic low-osmolar contrast medium iopromide (Ultravist 370, 370 mg/mL iodine), purchased from Bayer HealthCare LLC (Leverkusen, Germany), was used as the iodinated radiographic contrast agent. Furosemide was bought from Glpbio Technology (CA, USA, Cat. No.: GC12970). Human renal tubular epithelial cell lines, HK-2 cells, were bought from Procell Life Science & Technology Co. (Wuhan, China). The RNA isolation reagent Trizol and cDNA synthesis kit were bought from TaKaRa Bio. (Tokyo, Japan). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum and PBS buffer were purchased from Gibco Life Technologies (NY, USA). The cell counting kit-8 (CCK-8) test solution was bought from Dojindo Lab. (Japan). The TUNEL assay was purchased from Beyotime Bio. (Shanghai, China, Cat. No.: C1088).
Construction of in vivo CIAKI Model and Pretreatment with Icariin
All animal experiments were approved by Guangzhou Medical University and conducted in accordance with the US guidelines (NIH publication #85-23, revised in 1985). Mature male Sprague–Dawley rats, weighing 180–200 g, were chosen for this experiment. They were housed under controlled conditions of light (12 h/12 h light/dark cycle) and temperature (21–23°C) and allowed free access to standard laboratory diet and water. Rats were divided into three groups, while there are eight rats in each group: (1) control group: rats received saline solution at each time point; (2) CIAKI group: rats established CIAKI protocol; (3) CIAKI + icariin group: before establishment of CIAKI protocol, rats received icariin (100 mg/kg, in saline with a concentration of 1%) intragastrically once daily for three consecutive days. The dosage of icariin was determined according to previous studies.17,18 CIAKI protocol was established based on previous studies.19 Briefly, furosemide was administrated intragastrically at 200 mg/kg and dehydration for 6h before the contrast medium was injected at 15mL/kg by tail vein. After contrast administration, rats were allowed free access to water. Twenty-four hours after CIAKI protocol establishment, rats were weighed and obtained blood samples from orbital vein. And then the rats were killed under anesthesia (chloral hydrate (400 mg/kg, i.p.)). The kidneys were separated from renal capsules and weighed in the equatorial plane; one kidney was dissected and kept in liquid nitrogen for high throughput sequencing analysis, and the other was fixed in phosphate-buffered 4% formalin and prepared for histological examination.
Evaluation of Biochemical Parameters
Mean serum Cr, blood urea nitrogen (BUN) and Cystatin C (Cys-C) concentrations were measured using automatic biochemistry analyzer at the Clinical Laboratory, The Third Affiliated Hospital of Guangzhou Medical University.
Histological Examination of Renal Tissues
Kidney sections of 4μm were obtained from paraffin-embedded samples and subjected to hematoxylin and eosin (HE) staining and Periodic Acid-Schiff (PAS) staining. For semiquantitative analysis of morphological changes, 10 high-magnification (×200) fields of the cortex and outer stripe of the outer medulla were randomly selected. The tubular injury was evaluated according to the following criteria: tubular dilatation and necrosis, brush border loss and proteinaceous casts. Damage was graded on a scale from 0 to 4: 0, no abnormalities; 1+, damage less than 25%; 2+, damage between 25 and 49%; 3+, damage between 50 and 74%; 4+, damage more than 75%. The average score of 10 fields was used for each kidney sample.20 TUNEL assay was performed on paraffin-embedded kidney tissue to present apoptotic DNA fragmentation. Counterstained with DAPI, the apoptotic cells (FITC positive) appeared brightly green in color after fluorescent staining. For quantitative analysis, TUNEL-positive cells were analyzed in six random high-magnification (×400) fields using the Particle Analyzer function in ImageJ.21
RNA Extraction and Qualification
Three rats from each group were randomly chosen for whole-genome sequencing. Total RNA from each kidney was extracted using TRIzol reagent (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions. One percent agarose gels were used for RNA degradation and contamination. RNA concentration was measured using a Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit with the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
RNA Sequencing
All sequencing programs were carried out by Biomarker Company (Beijing, China). In brief, gene expression profiling of genome-wide kidney tissues was performed using RNA deep sequencing. Moreover, library construction was performed following the manufacturer’s instructions (Illumina, San Diego, CA). Samples were sequenced on an Illumina HiSeq 2500 instrument.
Bioinformatics
Bioinformatic analysis was performed by R language (version 3.4.4). The raw read counts of genes were normalized, and then transcripts per million (TPM) was applied. Differentially expressed genes (DEGs) were analyzed by “DeSeq2”, while absolute value of fold change (FC) ≥ 1.5 and adjusted p < 0.05 was considered significant. Then, the “EnhancedVolcano” and “pheatmap” packages were used to draw volcano maps and heatmaps. Gene Ontology (GO) analysis of differentially expressed (DE) mRNAs was achieved by Metascape web server.6 Predicted target genes were filtered by miRDB (http://mirdb.org/), Targetscan (http://www.targetscan.org/), circBase (http://www.circbase.org) and Lncpedia (https://lncipedia.org) databases. The ceRNA network was constructed based on DEGs. The cross-linking relationship and network construction were performed by Cytoscape version 3.6.0. For mRNAs in the ceRNA network, WebGestalt online tool (http://www.webgestalt.org) were utilized for function annotation, while another online tool String22 were used for protein–protein interaction (PPI) prediction. “Cytohubba” plug-in of Cytoscape was applied to identify key modules of the ceRNA network. “Ggalluvial” package was used for visualization of the ceRNA network as a Sankey plot. Based on the median level of Map3k14, the nine sequenced rats were divided into two groups. Then, Map3k14 associated GSEA analysis was carried out by WebGestalt online tool. “Corrplot” package was used for correlation analysis of renal function and key modules in the ceRNA network. The fitting curves of Cr and specific modules were exhibited by GraphPad Prism 9 software.
RNA Isolation and PCR Amplification
The total RNA was isolated from the kidney using Trizol. Reverse transcription into cDNA was performed using a PrimeScript II 1st Strand cDNA synthesis kit. The real-time quantitative PCR amplification was done by Power SYBR Green PCR Master Mix (Thermo Fisher). 50°C for 2 min, 95°C for 2 min, 40 cycles of 95°C for 15s, 60°C for 1min were set as the reaction conditions. The primers used were listed in Table S1. The relative expression levels were calculated using the 2−ΔΔCt methods.
In vitro Study of Icariin Protection in CM-Induced Renal Tubular Injury
HK-2 cells were preserved in DMEM with 10% fetal bovine serum under a condition of 37°C and 5% CO2 atmosphere. The culture medium was replaced with fresh medium every 2–3 days. The cells were expanded to new culture plates when about 80% confluent. HK-2 cells were exposed to CM at different concentrations for individual times to investigate its injury. In order to assess the effect of icariin against CM-induced injury, HK-2 cells were co-treated with icariin at different concentrations when exposed to CM.
The cell viability was detected by CCK-8 assay. The HK-2 cells were incubated in 96-well plate at a concentration of 1×104 cells/mL at 37°C. After indicated treatments, the cells were washed twice with PBS. Then the cells were incubated with 10μL CCK-8 test solution and 90 mL DMEM at 37 °C for 2 h. The optical density (OD) was measured by absorbance value at the 450nm wavelength using a microplate reader (Molecular Devices, USA). The mean of the OD of three wells in each group was used for calculation of cellular activity percentage, according to the following formula: cell viability (%) = (OD of treatment group/OD of control group) × 100%.
Statistical Analysis
All data were expressed as the mean ± SD. Differences between groups were determined by one-way analysis of variance (ANOVA) using SPSS 17.0 software (SPSS, Inc, Chicago, IL, USA) followed by the least significant difference (LSD) post hoc comparison test. Differences were considered statistically significant at p < 0.05.
Results
Icariin Protected Against CIAKI in in vivo and in vitro Models
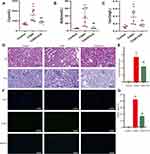
In Sprague–Dawley rats, iopromide caused significant elevation of serum Cr (Figure 1A), BUN (Figure 1B) and Cys-C (Figure 1C) levels. Prominent morphological changes, such as tubular dilatation and necrosis, proteinaceous casts and interstitial edema, were also observed in iopromide treated rat kidneys (Figure 1D and E). However, icariin pretreatment exhibited remarkable renal protection in CIAKI rats, as it alleviated serum Cr, BUN and Cys-C expressions (Figure 1A–C) and ameliorated morphological injury (Figure 1D and E). Moreover, icariin could dampen iopromide-induced renal apoptosis, presented as decreased TUNEL-positive renal tubular cells (Figure 1F and G).
In HK-2 cells, iopromide caused dose (Figure 2A) and time (Figure 2B) dependent injury. As iopromide with a concentration of 150 mg I/mL treating for 9 h could induce nearly 40% cell death, it was set as the CM-induced renal tubular injury model. Next, we tested the effect of icariin on cell viability alone. As shown in Figure 2C, icariin did not decrease cell viability in the concentration lower than 100 μM, which was then used for the following investigation. Further, we explored the effect of icariin in CM-induced renal tubular injury model. In Figure 2D, icariin could remarkably attenuate CM-induced renal tubular cell death in the concentration from 1 to 50 μM. The above data indicate a prominent protective effect of icariin against CIAKI in both in vivo and in vitro models.
Identification of DEGs in Icariin Pretreated CIAKI Rats
Next, we randomly chose three rats from each group, and carried out next-generation sequencing to the kidney tissues. Overview of gene expression in these nine rat kidneys is shown in Figure S1. Totally, there are 4348 DE mRNAs (Figure 3A), 149 DE miRNAs (Figure 3D), 1082 DE lncRNAs (Figure 3G) and 19 DE circRNAs (Figure 3J) between control and CIAKI group. Moreover, there are 3975 DE mRNAs (Figure 3B), 128 DE miRNAs (Figure 3E), 989 DE lncRNAs (Figure 3H) and 16 DE circRNAs (Figure 3K) between CIAKI and CIAKI+icariin group. The distribution of DEGs was also exhibited by circos plots (Figure S2A and S2C) and pie charts (Figure S2B and S2D). More details of DEGs were displayed in Tables 1–4. To further investigate genes attributed to icariin protection against CIAKI, we looked for DEGs with similar expression trends in control group and CIAKI+icariin group. Finally, we focused on 3112 mRNAs (Figure 3C), 62 miRNAs (Figure 3F), 396 lncRNAs (Figure 3I) and 5 circRNAs (Figure 3L).
 |
Table 1 Top 20 Differentially Expressed mRNAs |
 |
Table 2 Top 20 Differentially Expressed microRNAs |
 |
Table 3 Top 20 Differentially Expressed lncRNAs |
 |
Table 4 Differentially Expressed circRNAs |
Function Annotation of DEGs in Icariin Pretreated CIAKI Rats
To clarify the biological function of DE mRNAs, gene ontology (GO) annotation was conducted by Metascape. In Figure 4A, top 20 enriched GO terms were presented by a heatmap. DEGs from CIAKI group and CIAKI+icariin group were both enriched in response to drug (GO: 0042493), inflammatory response (GO: 0006954), response to wounding (GO:0009611) and Nrf-2 signaling (WP2376). Top 100 enriched GO terms were also exhibited in Figure S3. To better display the correlation in biological function between CIAKI and CIAKI+icariin groups, enriched GO terms were clustered as a network further. Combing Figure 4B and C, overlapped GO terms enriched in inflammatory response, leukocyte migration, response to wounding and Nrf-2 signaling might be part of the underlying mechanism in icariin protection against CIAKI. Specific information of top enriched GO terms is demonstrated in Table 5.
 |
Table 5 Top 10 Enriched GO Terms of DEGs |
Construction of Icariin-Associated ceRNA Networks in CIAKI Rats
In order to figure out the interaction between protein-coding genes and ncRNAs in icariin protection against CIAKI, we tried to construct ceRNA networks using the selected 3112 mRNAs, 62 miRNAs, 396 lncRNAs and 5 circRNAs. By cytoscape, we set up a lncRNA-associated ceRNA network (Figure S4), including 417 edges and 389 nodes (93 lncRNAs, 15 miRNAs and 281 mRNAs). We also set up a circRNA-associated ceRNA network (Figure S5), including 277 edges and 264 nodes (4 circRNAs, 8 miRNAs and 252 mRNAs). Each ceRNA network was divided into two parts, one with up-regulated miRNAs in CIAKI+icariin group, the other with down-regulated miRNAs.
Enrichment Analysis and PPI Analysis of Icariin-Associated ceRNA Networks
Further, we focused on the potential molecular mechanism and pathways of ceRNA networks participating in icariin protection against CIAKI. Using the WebGestalt online tool, we found icariin-associated ceRNA networks were highly enriched in biological regulation and metabolic process among biological process (BP) categories, in membrane and nucleus among cellular component (CC) categories, as well as in protein binding and ion binding among molecular function (MF) categories (Figure S6A). We also determined top 20 enriched BP terms of the lncRNA-associated ceRNA network (Figure 5A) and circRNA-associated ceRNA network (Figure 5B). We found that mRNAs in the lncRNA-associated ceRNA network were positively correlated with ERK1 and ERK2 cascade, cell-cell adhesion and regulation of cellular response to stress, while negatively correlated with response to oxidative stress, response to interleukin-1 and ncRNA metabolic process. On the other hand, mRNAs in the circRNA-associated ceRNA network were positively correlated with Ras and cGMP-PKG signaling pathways, while negatively correlated with MAPK and NF-κB signaling pathways. Top 20 enriched CC (Figure S6B) and MF (Figure S6C) terms of icariin-associated ceRNA networks were also demonstrated.
Using an online tool String, we also investigated the protein–protein interaction of mRNAs in icariin-associated ceRNA networks (Figure 5C). Interestingly, Map3k14, a canonical NF-κB signaling inducing kinase, was involved in hub genes of the PPI network, as well as Utp15, Epha4 and Rps3.
Identification and Validation of Key Modules in Icariin-Associated ceRNA Networks
To find out key modules of the icariin associated ceRNA networks, the “Cytohubba” plug-in of Cytoscape software was applied. Combining seven methods in “Cytohubba” (betweenness, mcc, closeness, degree, epc, radiality and stress), we figured out a hub lncRNA-associated ceRNA network with 4 miRNAs (rno-miR-146b-5p, rno-miR-339-5p, rno-miR-345-3p and rno-miR-6328), 6 lncRNAs (MSTRG.34520.3, MSTRG.4074.10, MSTRG.51241.14, MSTRG.51241.15, MSTRG.69307.1 and MSTRG.71029.16) and 6 mRNAs (Acot1, Ly6i, Map3k14, Mettl2, Set and Tmem44). We also set up a hub circRNA-associated ceRNA network with 1 miRNA (rno-miR-339-5p), 2 circRNAs (10:13767446|13810899 and 3:177390074|177433646) and 8 mRNAs (Acot1, Cbwd1, Ly6i, Map3k14, Mettl2, Nyap1, Set and Utp20). An overview of the hub ceRNA network was also demonstrated by a Sankey plot (Figure 6A). Detail of each hub gene is presented in Tables 6 and 7.
 |
Table 6 Hub Nodes of lncRNA-Associated ceRNA Analysis |
 |
Table 7 Hub Nodes of circRNA-Associated ceRNA Analysis |
Further, we detected the expressions of above 9 mRNAs involved in the icariin-associated ceRNA network by PCR analysis. As shown in Figure 6B, CM induced significant elevation of Acot1, Map3k14 and Ly6i, which could be reversed by icariin pretreatment. Icariin pretreatment also attenuated CM-induced expressions of Mettl2 and Set.
From the hub ceRNA networks, we focused on the overlapped mRNA Map3k14. Based on the level of Map3k14 in the nine sequenced rat kidneys, we carried out GSEA analysis. We could see from Figure S7, mRNA metabolic process (NES = 2.332, p < 0.001), ncRNA metabolic process (NES = 2.996, p < 0.001) and apoptotic signaling pathway (NES = 2.257, p < 0.001) were positively enriched.
Relationship of Icariin-Associated ceRNA Networks and Renal Function
Finally, we tried to inspect the relationship between the key modules of ceRNA networks and renal function. Using an R package “corrplot”, Figure 7A and Figure S8 were drawn to demonstrate the correlation of 21 key modules and renal function (Cr, BUN and Cys-C). Three biomarkers of renal function showed similar trends of correlation with the key modules. They were negatively correlated with one mRNA (Tmem44), one miRNA (rno-miR-339-5p) and one lncRNA (MSTRG.4074.10). On the other hand, they were positively correlated with eight mRNAs (Acot1, Cbwd1, Ly6i, Map3k14, Mettl2, Nyap1, Set and Utp20), two miRNAs (rno-miR-345-3p and rno-miR-6328), one lncRNA (MSTRG.69307.1) and one circRNA (10:13767446|13810899).
More specifically, fitting curves of Cr and key modules were exhibited. Acot1 (Figure 7B, p < 0.001), Cbwd1 (Figure 7C, p < 0.001), Map3k14 (Figure 7D, p = 0.02), Nyap1 (Figure 7E, p < 0.001), rno-miR-6328 (Figure 7H, p = 0.001) and lncRNA MSTRG.69307.1 (Figure 7J, p = 0.02) were positively correlated with Cr level. Meanwhile, Tmem44 (Figure 7F, p = 0.007), rno-miR-339-5p (Figure 7G, p = 0.003) and lncRNA MSTRG.4074.10 (Figure 7I, p = 0.004) were negatively correlated with Cr level.
Discussion
In the present study, we reported the protective effect of icariin against CIAKI. We also demonstrated the expression profiles and function annotation of mRNAs, miRNAs, lncRNAs and circRNAs in icariin pretreated CIAKI rat kidneys. Further, we figured out an icariin-associated ceRNA network and evaluated its relationship with renal function.
The pathophysiological mechanisms of CIAKI have not been thoroughly elucidated, but direct nephrotoxic effect of CM and disturbance of renal blood flow have been generally recognized.5,23 CM are toxic to the tubular epithelium, leading to inflammation and apoptosis. With progressed cellular injury, tubular epithelial cells detach from the basement membrane and cause luminal obstruction, and finally cause a decrease of glomerular filtration rate.5 Disturbed renal blood flow can cause arteriolar vasoconstriction and microvascular thrombosis, which enhance the renal tubular injury. Therefore, preventive strategies targeting the above pathogenesis may be effective. In this study, icariin significantly protected against CM-induced renal tubular apoptosis, as a profound decrease in numbers of TUNEL-positive tubular cells was observed. Function annotation of DEGs showed an enrichment of inflammatory response, regulation of cell adhesion, leukocyte migration and blood vessel development in icariin pretreated CIAKI rats. Similarly, Xie et al reported a protection of icariin against inflammatory response, apoptosis and vascular permeability in a sepsis-induced AKI mouse model.24 In another unilateral ureteral obstruction mouse model, icariin also mitigated several proinflammatory factors, such as NF-κB, cyclooxygenase-2 and interleukin 1β.10 Thus, the universal anti-inflammation and anti-apoptosis effect of icariin may contribute to its remarkable protection in CIAKI model, as the improved renal function and alleviated histological injury were demonstrated in our study. To our knowledge, this is the first report about the protective effect of icariin against CIAKI.
Then, we focused on the potential role of ncRNAs in icariin protection against CIAKI. ncRNAs, as important epigenetic modulators, can induce changes to a phenotype without influencing a genotype. They are potentially reversible and can be affected by environmental factors, age and disease state. Emerging evidence suggests a crucial role of ncRNAs in various kidney diseases, such as AKI, CKD, diabetic kidney disease and renal carcinoma. For example, Lorenzen et al demonstrated elevation of three novel circRNAs in blood of patients with AKI, among which circR-126 might play as a predictor of mortality.25 LncRNA H19, elevated in kidney biopsies of patients with AKI and in murine ischemic kidney tissues was shown to attenuate ischemic AKI by sponging miRNA-30a-5p.26 In the present study, we found 128 DE miRNAs, 989 DE lncRNAs and 16 DE circRNAs between CIAKI and CIAKI+icariin group. Particularly, rno-miR-339-5p was up-regulated in CIAKI+icariin group, while rno-miR-146b-5p and rno-miR-345-3p were down-regulated. MiR-339-5p was proven to be associated with risk of end stage kidney disease in diabetic patients.27 In glomerular mesangial cells, miR-146b-5p attenuated the inflammatory response by inhibiting Toll-like receptor 4 pathway in lupus nephritis.28 MiR-345-3p also presented significant pathological relevance in cardiorenal syndrome.29 Focused on so many popular ncRNAs, we were also curious about the underlying mechanism and interaction of these ncRNAs in icariin protection against CIAKI.
The foundation of ceRNA hypothesis is that both coding and non-coding endogenous RNA molecules sharing miRNA response elements (MREs) can competitively prevent the action of miRNAs on their mRNA targets, thereby regulate gene expression in trans.30 A variety of experiments have proven the crucial role of ceRNA networks in AKI. For example, a ceRNA axis involving lncRNA TCONS_00016233, miR-22-3p and AIFM1 contributed to TLR4/MAPK signal pathway and enhanced septic AKI.31 Bao et al constructed a ceRNA network including 22 lncRNAs, 5 miRNAs and 37 mRNAs in a CIAKI rat model, which was functionally enriched in cell adhesion molecules (rno04514), intracellular signal transduction (GO:0035556) and transmembrane transporter activity (GO:0022857).16 In another study focused on CIAKI, Cheng et al demonstrated a ceRNA network involving 38 circRNAs, 12 lncRNAs, 13 miRNAs and 127 mRNAs.32 Further function annotation of that study presented an enrichment in TNF signaling pathway (rno04668), RNA splicing and nucleoside binding. In our study, based on the DEGs between CIAKI group and CIAKI+icariin group, we constructed a lncRNA-associated ceRNA network (including 93 lncRNAs, 15 miRNAs and 281 mRNAs) and a circRNA-associated ceRNA network (including 4 circRNAs, 8 miRNAs and 252 mRNAs). Similarly, mRNAs in the ceRNA networks were remarkably enriched in ncRNA metabolic process, cell-cell adhesion and several canonical signal pathways (MAPK signaling, NF-κB signaling, calcium signaling, ERK1 and ERK2 cascade). Our results indicated that the function of ceRNA network in icariin protection against CIAKI might consist with that in CIAKI pathogenesis, or even with that in other AKI models. Moreover, we identified 21 key modules (involving 6 lncRNAs, 2 circRNAs, 4 miRNAs and 9 mRNAs) in these ceRNA networks, which might be highly correlated with the protective effect of icariin in CIAKI. Interestingly, rno-miR-339-5p, rno-miR-345-3p and rno-miR-146b-5p were among top 5 hub genes in the 22 key modules. Additionally, rno-miR-339-5p showed negative correlation with rats’ Cr level, while rno-miR-345-3p and rno-miR-146b-5p showed positive correlation. As mentioned above, these three miRNAs all presented relevance with pathogenesis and prognosis in other kidney diseases, which prompted a crucial role of ceRNA networks in icariin protection against CIAKI.
Map3k14, an NF-κB inducing kinase, showed up as a core gene in the mechanism of icariin protection against CIAKI in the present study. It was among the hub genes both in the ceRNA network and in the PPI network, and it was associated with deteriorating renal function. ncRNA and mRNA metabolism, as well as apoptotic signaling, were all highly correlated with Map3k14 expression pattern. Consistently, Map3k14 has been proven to promote inflammation and cell death in acute folate nephropathy.33 Map3k14 overexpression increased inflammatory cytokines such as IL-6, IL-8 and MCP-1 in cultured human proximal tubular cells.34 Mechanistically, Map3k14 functions by regulation of both canonical and non-canonical NF-κB pathways, as well as NF-κB-independent actions (such as regulation of transcription factor activity and epigenetic mechanism).35 These might account for the underlying mechanism of icariin protection in CIAKI partially, as the icariin-associated ceRNA network was also enriched in ncRNA metabolism and NF-κB signaling.
Despite these thought-provoking results, there are certain limitations of our study. First, we selected three rat kidneys in each group for sequencing analysis. Further experiments with a larger number of samples and a greater variety of tissue, including more precise renal medulla and cortex, are needed for sequencing analysis. Second, we only focused on the kidney injury induced by one type of CM—iopromide. Therefore, we will discuss the effect of icariin in CIAKI models induced by different CM. Third, we did not verify ceRNA network in the in vitro CIAKI models, which should be covered in the future. Finally, we did not involve human samples in the present study. We will collect patients’ blood and kidney samples for further investigation to increase persuasion of the study.
Conclusion
In the current study, we reported, for the first time, the protective effect of icariin in CIAKI via both in vivo and in vitro studies. The ceRNA network based on high-throughput sequencing revealed that Acot1, Cbwd1, Ly6i, Map3k14, Mettl2, Nyap1, Set, Tmem44 and Utp20 were crucial genes in icariin action. The lncRNA/circRNA-miRNA-mRNA interaction further prompted that ERK1 and ERK2 cascade, MAPK signaling and NF-κB signaling might take part in the icariin protection against CIAKI. The above results may shed a light on succeeding studies about the preventive strategies of CIAKI.
Abbreviations
AKI, acute kidney injury; ANOVA, analysis of variance; BUN, blood urea nitrogen; CC, cellular component; CCK-8, cell counting kit-8; ceRNA, competing endogenous RNA; CIAKI, contrast induced acute kidney injury; circRNA, circular RNA; CM, contrast medium; Cr, creatinine; Cys-C, Cystatin C; DE, differentially expressed; DEG, differentially expressed genes; FC, fold change; GO, gene ontology; HE, hematoxylin and eosin; lncRNA, long non-coding RNA; LSD, least significant difference; MF, molecular function; miRNA, microRNA; MRE, miRNA response element; ncRNA, non-coding RNA; PAS, Periodic Acid-Schiff; PPI, protein–protein interaction; TPM, transcripts per million.
Data Sharing Statement
The datasets generated and analyzed during current study are available in the GEO repository [GSE189881].
Ethics Approval and Consent to Participate
All animal experiments were approved by Guangzhou Medical University and conducted in accordance with the US guidelines (NIH publication #85-23, revised in 1985).
Funding
This work was supported by National Natural Science Foundation of China (grant numbers 82000355 and 81902831), Guangdong Provincial Natural Science Foundation (grant number 2021A1515111114), Guangzhou Science and Technology Program (grant numbers 202102080037 and 202102020831), Guangzhou Traditional Chinese Medicine and Integrated Medicine Technology Program (grant number 20212A11023) and Scientific research funding of Traditional Chinese Medicine Bureau of Guangdong Province (grant number 20221246).
Disclosure
The authors declare that they have no competing interests.
References
1. Azzalini L, Spagnoli V, Ly HQ. Contrast-induced nephropathy: from pathophysiology to preventive strategies. Can J Cardiol. 2016;32(2):247–255. doi:10.1016/j.cjca.2015.05.013
2. Kellum JA, Lameire N. Diagnosis, evaluation, and management of acute kidney injury: a KDIGO summary (Part 1). Critical Care. 2013;17(1):204. doi:10.1186/cc11454
3. Shema L, Ore L, Geron R, Kristal B. Contrast-induced nephropathy among Israeli hospitalized patients: incidence, risk factors, length of stay and mortality. Isr Med Assoc J. 2009;11(8):460–464.
4. Yan L, Jiaqiong L, Yue G, et al. Atorvastatin protects against contrast-induced acute kidney injury via upregulation of endogenous hydrogen sulfide. Ren Fail. 2020;42(1):270–281. doi:10.1080/0886022x.2020.1740098
5. Mehran R, Dangas GD, Weisbord SD. Contrast-associated acute kidney injury. N Engl J Med. 2019;380(22):2146–2155. doi:10.1056/NEJMra1805256
6. Nijssen EC, Rennenberg RJ, Nelemans PJ, et al. Prophylactic hydration to protect renal function from intravascular iodinated contrast material in patients at high risk of contrast-induced nephropathy (AMACING): a prospective, randomised, Phase 3, controlled, open-label, non-inferiority trial. Lancet. 2017;389(10076):1312–1322. doi:10.1016/s0140-6736(17)30057-0
7. Sze SC, Tong Y, Ng TB, Cheng CL, Cheung HP. Herba Epimedii: anti-oxidative properties and its medical implications. Molecules. 2010;15(11):7861–7870. doi:10.3390/molecules15117861
8. Li C, Li Q, Mei Q, Lu T. Pharmacological effects and pharmacokinetic properties of icariin, the major bioactive component in Herba Epimedii. Life Sci. 2015;126:57–68. doi:10.1016/j.lfs.2015.01.006
9. Zhou YD, Hou JG, Yang G, et al. Icariin ameliorates cisplatin-induced cytotoxicity in human embryonic kidney 293 cells by suppressing ROS-mediated PI3K/Akt pathway. Biomed Pharmacother. 2019;109:2309–2317. doi:10.1016/j.biopha.2018.11.108
10. Chen HA, Chen CM, Guan SS, Chiang CK, Wu CT, Liu SH. The antifibrotic and anti-inflammatory effects of icariin on the kidney in a unilateral ureteral obstruction mouse model. Phytomedicine. 2019;59:152917. doi:10.1016/j.phymed.2019.152917
11. Jia Z, Wang K, Zhang Y, et al. Icariin ameliorates diabetic renal tubulointerstitial fibrosis by restoring autophagy via regulation of the miR-192-5p/GLP-1R pathway. Front Pharmacol. 2021;12:720387. doi:10.3389/fphar.2021.720387
12. Wang M, Wang L, Zhou Y, Feng X, Ye C, Wang C. Icariin attenuates renal fibrosis in chronic kidney disease by inhibiting interleukin-1β/transforming growth factor-β-mediated activation of renal fibroblasts. Phytother Res. 2021;35:6204–6215. doi:10.1002/ptr.7256
13. Qi MY, He YH, Cheng Y, et al. Icariin ameliorates streptozocin-induced diabetic nephropathy through suppressing the TLR4/NF-κB signal pathway. Food Funct. 2021;12(3):1241–1251. doi:10.1039/d0fo02335c
14. Haddad G, Kölling M, Wegmann UA, et al. Renal AAV2-mediated overexpression of long non-coding RNA H19 attenuates ischemic acute kidney injury through sponging of microRNA-30a-5p. J Am Soc Nephrol. 2021;32(2):323–341. doi:10.1681/asn.2020060775
15. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
16. Bao W, Xiao Z, Wang Z, Liu D, Tan P, Huang M. Comprehensive analysis of the long non-coding RNA expression profile and functional roles in a contrast-induced acute kidney injury rat model. Exp Ther Med. 2021;22(1):739. doi:10.3892/etm.2021.10171
17. Zhang X, Chen Y, Zhang C, et al. Effects of icariin on the fracture healing in young and old rats and its mechanism. Pharm Biol. 2021;59(1):1245–1255. doi:10.1080/13880209.2021.1972121
18. Zhang W, Yuan W, Xu N, Li J, Chang W. Icariin improves acute kidney injury and proteinuria in a rat model of pregnancy‑induced hypertension. Mol Med Rep. 2017;16(5):7398–7404. doi:10.3892/mmr.2017.7513
19. Liu Y, Liu B, Liu Y, et al. MicroRNA expression profile by next-generation sequencing in a novel rat model of contrast-induced acute kidney injury. Ann Transl Med. 2019;7(8):178. doi:10.21037/atm.2019.04.44
20. Weidemann A, Bernhardt WM, Klanke B, et al. HIF activation protects from acute kidney injury. J Am Soc Nephrol. 2008;19(3):486–494. doi:10.1681/asn.2007040419
21. Li J, Xu S, Zhu JB, et al. Pretreatment with cholecalciferol alleviates renal cellular stress response during ischemia/reperfusion-induced acute kidney injury. Oxid Med Cell Longev. 2019;2019:1897316. doi:10.1155/2019/1897316
22. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d13. doi:10.1093/nar/gky1131
23. Tan X, Zheng X, Huang Z, Lin J, Xie C, Lin Y. Involvement of S100A8/A9-TLR4-NLRP3 inflammasome pathway in contrast-induced acute kidney injury. Cell Physiol Biochem. 2017;43(1):209–222. doi:10.1159/000480340
24. Xie C, Liu L, Wang Z, et al. Icariin improves sepsis-induced mortality and acute kidney injury. Pharmacology. 2018;102(3–4):196–205. doi:10.1159/000487955
25. Kölling M, Seeger H, Haddad G, et al. The circular RNA ciRs-126 predicts survival in critically ill patients with acute kidney injury. Kidney Int Rep. 2018;3(5):1144–1152. doi:10.1016/j.ekir.2018.05.012
26. He Y, Lang X, Cheng D, Zhang T, Yang Z, Xiong R. miR‑30a‑5p inhibits hypoxia/reoxygenation‑induced oxidative stress and apoptosis in HK‑2 renal tubular epithelial cells by targeting glutamate dehydrogenase 1 (GLUD1). Oncol Rep. 2020;44(4):1539–1549. doi:10.3892/or.2020.7718
27. Satake E, Saulnier PJ, Kobayashi H, et al. Comprehensive search for novel circulating miRNAs and axon guidance pathway proteins associated with risk of ESKD in diabetes. J Am Soc Nephrol. 2021;32(9):2331–2351. doi:10.1681/asn.2021010105
28. Sheng ZX, Yao H, Cai ZY. The role of miR-146b-5p in TLR4 pathway of glomerular mesangial cells with lupus nephritis. Eur Rev Med Pharmacol Sci. 2018;22(6):1737–1743. doi:10.26355/eurrev_201803_14589
29. Ishrat R, Ahmed MM, Tazyeen S, et al. In silico integrative approach revealed key MicroRNAs and associated target genes in cardiorenal syndrome. Bioinform Biol Insights. 2021;15:11779322211027396. doi:10.1177/11779322211027396
30. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
31. Zhang P, Yi L, Qu S, et al. The biomarker TCONS_00016233 drives septic AKI by targeting the miR-22-3p/AIFM1 signaling axis. Mol Ther Nucleic Acids. 2020;19:1027–1042. doi:10.1016/j.omtn.2019.12.037
32. Cheng W, Li XW, Xiao YQ, Duan SB. Non-coding RNA-associated ceRNA networks in a new contrast-induced acute kidney injury rat model. Mol Ther Nucleic Acids. 2019;17:102–112. doi:10.1016/j.omtn.2019.05.011
33. Ortiz A, Husi H, Gonzalez-Lafuente L, et al. Mitogen-activated protein kinase 14 promotes AKI. J Am Soc Nephrol. 2017;28(3):823–836. doi:10.1681/asn.2015080898
34. Zhao Y, Banerjee S, LeJeune WS, Choudhary S, Tilton RG. NF-κB-inducing kinase increases renal tubule epithelial inflammation associated with diabetes. Exp Diabetes Res. 2011;2011:192564. doi:10.1155/2011/192564
35. Valiño-Rivas L, Vaquero JJ, Sucunza D, et al. NIK as a druggable mediator of tissue injury. Trends Mol Med. 2019;25(4):341–360. doi:10.1016/j.molmed.2019.02.005
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.