Back to Journals » International Journal of Nanomedicine » Volume 19
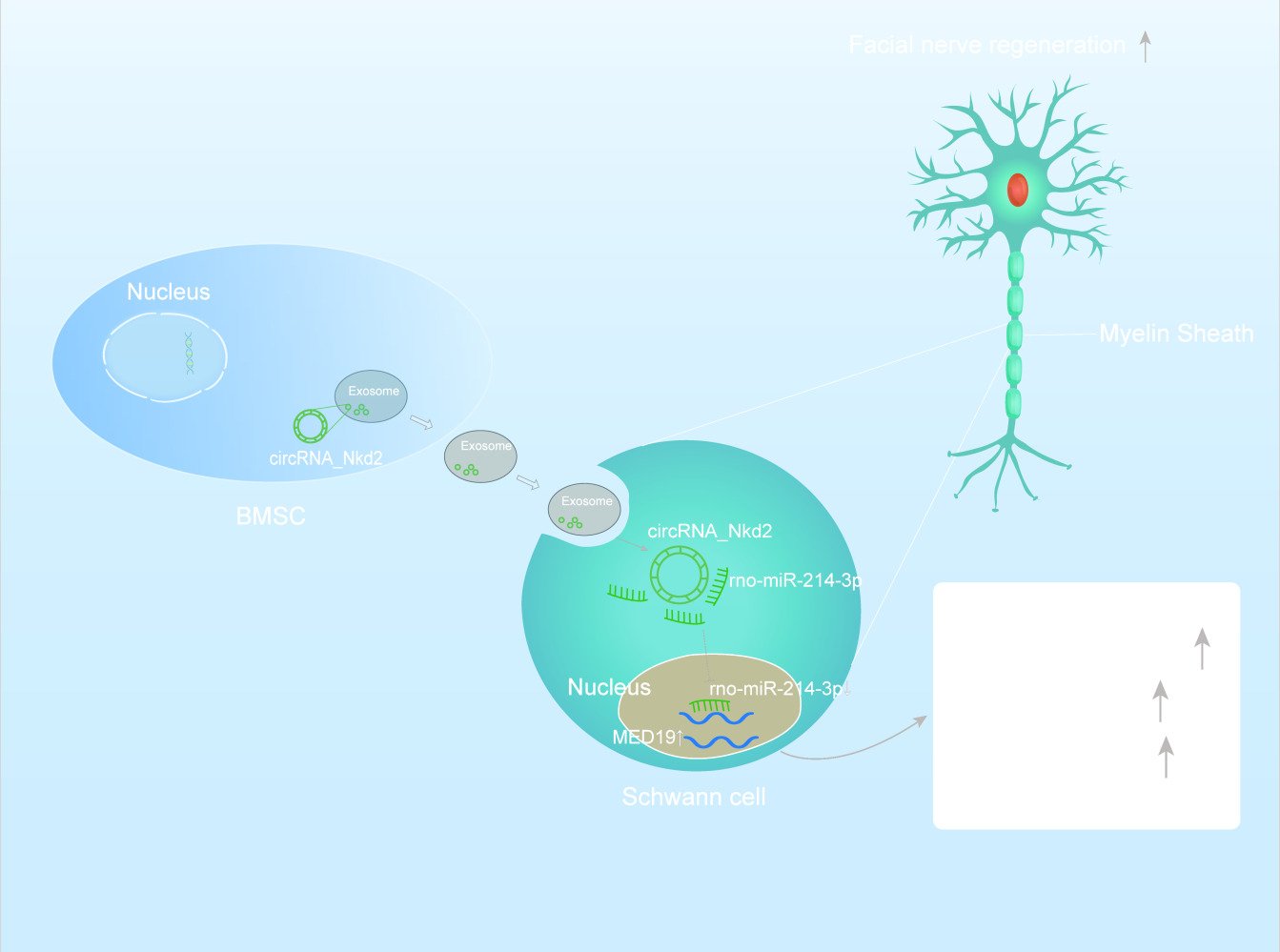
Hypoxic Bone Mesenchymal Stem Cell-Derived Exosomes Direct Schwann Cells Proliferation, Migration, and Paracrine to Accelerate Facial Nerve Regeneration via circRNA_Nkd2/miR-214-3p/MED19 Axis
Authors Wang H, Zhao H, Chen Z, Cai X, Wang X, Zhou P, Tang Y, Ying T, Zhang X, Shen Y, Wang B, Zhu W, Zhu J, Wang X , Li S
Received 26 October 2023
Accepted for publication 27 January 2024
Published 12 February 2024 Volume 2024:19 Pages 1409—1429
DOI https://doi.org/10.2147/IJN.S443036
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xing Zhang
Haopeng Wang,1,* Hua Zhao,1,* Zheng Chen,1,* Xiaomin Cai,1 Xuhui Wang,1 Ping Zhou,1 Yinda Tang,1 Tingting Ying,1 Xin Zhang,1 Yiman Shen,1 Baimiao Wang,1 Wanchun Zhu,1 Jin Zhu,1 Xinjun Wang,2 Shiting Li1
1Department of Neurosurgery, Xinhua Hospital Affiliated to Shanghai JiaoTong University School of Medicine, Shanghai, 200092, People’s Republic of China; 2Department of Neurosurgery, The Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, 450052, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shiting Li, Department of Neurosurgery, Xinhua Hospital Affiliated to Shanghai JiaoTong University School of Medicine, Shanghai, 200092, People’s Republic of China, Email [email protected] Xinjun Wang, Department of Neurosurgery, The Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, 450052, People’s Republic of China, Email [email protected]
Background: Facial nerves have the potential for regeneration following injury, but this process is often challenging and slow. Schwann cells (SCs) are pivotal in this process. Bone mesenchymal stem cells (BMSC)-derived exosomes promote tissue repair through paracrine action, with hypoxic preconditioning enhancing their effects. The main purpose of this study was to determine whether hypoxia-preconditioned BMSC-derived exosomes (Hypo-Exos) exhibit a greater therapeutic effect on facial nerve repair/regeneration and reveal the mechanism.
Methods: CCK-8, EdU, Transwell, and ELISA assays were used to evaluate the functions of Hypo-Exos in SCs. Histological analysis and Vibrissae Movements (VMs) recovery were used to evaluate the therapeutic effects of Hypo-Exos in rat model. circRNA array was used to identify the significantly differentially expressed exosomal circRNAs between normoxia-preconditioned BMSC-derived exosomes (Nor-Exos) and Hypo-Exos. miRDB, TargetScan, double luciferase assay, qRT-PCR and WB were used to predict and identify potential exosomal cirRNA_Nkd2-complementary miRNAs and its target gene. The function of exosomal circRNA_Nkd2 in facial nerve repair/regeneration was evaluated by cell and animal experiments.
Results: This study confirmed that Hypo-Exos more effectively promote SCs proliferation, migration, and paracrine function, accelerating facial nerve repair following facial nerve injury (FNI) compared with Nor-Exos. Furthermore, circRNA analysis identified significant enrichment of circRNA_Nkd2 in Hypo-Exos compared with Nor-Exos. Exosomal circRNA_Nkd2 positively regulates mediator complex subunit 19 (MED19) expression by sponging rno-miR-214-3p.
Conclusion: Our results demonstrated a mechanism by which Hypo-Exos enhanced SCs proliferation, migration, and paracrine function and facial nerve repair and regeneration following FNI through the circRNA_Nkd2/miR-214-3p/Med19 axis. Hypoxic preconditioning is an effective and promising method for optimizing the therapeutic action of BMSC-derived exosomes in FNI.
Keywords: hypoxic, BMSCs, exosomes, Schwann cells, facial nerve injury
Graphical Abstract:
Introduction
Facial nerve injury is a peripheral nerve disorder, affecting millions of people annually worldwide, resulting in heave societal and economic burdens.1,2 Traditional treatments for restoring facial nerve function include medication, physical therapy, rehabilitation training, and surgery. Despite the array of clinical interventions available, the level of functional recovery post-FNI remains suboptimal.3 Therefore, elucidating the pathogenic mechanisms is crucial to identify effective strategies for enhancing facial nerve function recovery.
The facial nerve, a peripheral nerve, relies heavily on Schwann cells (SCs) during the repair process post-injury. SCs actions encompass migration, proliferation, differentiation, and paracrine effects in injured regions, significantly influencing the ultimate restoration of facial nerve function.4,5 Consequently, investigating methods to boost SCs migration, proliferation, differentiation, and paracrine effects in the injured area and to harness their intrinsic potential in nerve function repair by augmenting SCs numbers and density.
Research has underscored the therapeutic potential of bone mesenchymal stem cells (BMSCs) transplantation in tissue repair.6 BMSCs primarily achieve these benefits by secreting neurotrophins, promoting SCs migration and proliferation, and enhancing peripheral nerve regeneration through neuronal or glial differentiation.7 However, uncertainties persist concerning the safety of stem cell transplantation therapy, including risks such as immune rejection, cellular dedifferentiation, and tumor formation, limiting the clinical applicability of BMSCs transplantation.8,9 Recent investigations have highlighted that the therapeutic actions of BMSCs are primarily mediated through their paracrine functions, with that exosomes emerging as key players in this process.10
Exosomes are extracellular vesicles generated through endocytosis, released into the extracellular environment by fusing with the plasma membranes of various cell types. These exosomes contain proteins, mRNAs, microRNAs (miRNAs), circular RNAs (circRNAs), and other noncoding RNAs in their lumen. These exosomal RNAs can be taken up by nearby or distant cells via exosome circulation, influencing the functions of recipient cells, underscoring the growing interest in exosomes.11 Stem cell-secreted exosomes share biological functions akin to those of stem cells but with reduced adverse effects.12 For example, mesenchymal stem cell (MSCs)-derived exosomes often exhibit properties similar to those of MSCs, such as promoting tissue healing and mitigating inflammatory responses and fibrosis. The therapeutic potential of exosomes has drawn increasing attention in the context of tissue healing and nerve repair.13,14 Chen et al demonstrated that exosomes derived from human adipose-derived stem cells effectively promote peripheral nerve regeneration by optimizing SCs function.15
Oxygen concentration significantly influences BMSCs proliferation and differentiation. Conventional cell-culture practices expose BMSCs to normoxic conditions (21%), but in vivo, BMSCs typically experience hypoxia within tissues.16 Exosomes produced by hypoxia-pretreated BMSCs have demonstrated enhanced therapeutic effects, containing a variety of proteins and miRNAs.17 For example, recent research revealed that hypoxic BMSC-derived exosomes mitigate ulcerative colitis injury by reducing reactive oxygen species accumulation and DNA damage in intestinal epithelial cells through HIF-1α.18
Nonetheless, the potential contribution of BMSCs-derived exosomes under hypoxic conditions to the treatment of facial nerve injuries remains uncharted territory. In this study, we hypothesize that hypoxia-primed exosomes promote SCs proliferation, migration, and paracrine function through the circRNANkd2/miR-214-3p/mediator complex subunit 19 (MED19) axis, ultimately facilitating FNI repair.
Materials and Methods
Cell Culture and Hypoxia Treatment
The RSC96 cell line was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell was maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and 100 U/mL penicillin and 100 μg/mL streptomycin (1% P/S) (Gibco, USA) in a humidified incubator with 5% CO2 at 37 °C, and was passaged three times a week.
Primary Schwann cells (pSCs) was established and maintained from our lab as previously described.19 The cells were incubated with MEM/F12 (Gibco, USA) supplemented with 10% FBS in a humidified incubator with 5% CO2 at 37 °C. pSCs were identified at passage 3 (P3) for subsequent experiments.
Primary bone marrow mesenchymal stem cells (BMSCs) were extracted from bilateral femurs of male SD rats(70–100 g). In brief, after dissecting thigh bones under sterile conditions, DMEM (Cyagen, China) was added to flush the marrow cavities to obtain the suspension. Subsequently, gentle pipetting was performed to separate the cells. Later, BMSCs were obtained by centrifugation at 1000 rpm for 5 min. Next, BMSCs were cultivated within DMEM supplemented with 1% P/S and 10% FBS (Cyagen, China) in a humidified incubator with 5% CO2 at 37 °C. The cells were passaged twice a week and were used at the third passage. Flow cytometry (Beckman Coulter, USA) was performed to analyze the BMSCs positive markers (CD44, CD90, CD29, CD73) and negative markers (CD34, CD11b/c, CD45) (RAXMX-09011, Cyagen, China).
Exosome Isolation and Identification
BMSCs were cultured at normoxic condition (37 °C, 5% CO2, 21% O2) or at hypoxic condition (1% O2, 94% N2 and 5% CO2) using an oxygen control incubator (Thermo Fisher Scientific, USA) in exosome-depleted FBS conditioned (Cyagen, China) medium for 48 h. And then, the culture supernatants were subjected to sequential centrifugation steps at 300 ×g for 10 min, followed by 2000 ×g for 20 min and then at 10,000 ×g for 30 min to effectively remove cells and debris. The resulting supernatants were further processed through ultracentrifugation at 100,000 ×g for 70 min at a temperature of 4°C to pellet the exosomes. Finally, the exosome pellets were thoroughly washed with PBS and subsequently recovered by centrifugation at 100,000 ×g for another duration of 70 min.
Nanoparticle tracking analysis (NTA) (Nanosight Ltd, CA) was applied to analyze the distribution of vesicle diameters from the exosomes. The morphology of the acquired exosomes under normoxia and hypoxia condition was observed under a transmission electron microscope (TEM) (Hitachi, HT-7700, Japan). Western blotting was performed to determine the presence of specific exosomal surface markers, such as TSG101, CD63, and CD81. Bicinchoninic acid protein assay (BCA, Beyotime, China) was used to determine exosome protein concentration.
Exosome Internalization
To determine exosomes were internalized by RSC96 and pSCs, exosomes were labeled with the Dil solution (Molecular Probes, Beyotime, China) according to the manufacturer’s protocol. Briefly, 10µM Dil solution was added to PBS containing exosomes and incubated 30 min at 37 °C, and then incubated with RSC96 and pSCs at 37 °C. After 24 h, the cells were fixed in 4% paraformaldehyde for 15 min and subsequently blocked with PBS containing 1% BSA for a duration of 20–30 min. Following this, they were washed three times with PBS. The nuclei were dyed with DAPI (0.5 µg/mL, Beyotime, China) for 10 min, and the specimens were observed by confocal microscopy (Leica, Germany).
Cell Proliferation and Analysis
Cell (RSC96 and pSCs) proliferation was assessed by cell counting kit-8 solution (CCK8) (C0037, Beyotime, China) and Ethynyldeoxyuridine (EdU) retention assay (C0075S, Beyotime, China). For CCK8 assay, about 2×103 cells/well were seeded in 96-well plate. Then 10 μL of CCK8 solution in fresh culture medium was added to each well and incubated for 1 h at 37 °C in dark area. The microplate reader was used to test the optical density (OD) value of each well.
The EdU-555 Cell Proliferation Kit was also used to measure cell proliferation according to manufacturer’s instructions. Briefly, 2×104 cells were seeded in 24-well culture dishes and co-cultured with exosome for 48 h. EdU reagent (10 μM) was added to culture media for 2 h or 12 h to label RSC96 cell lines and pSCs, respectively. After being washed by PBS and fixed in 4% paraformaldehyde, the cells were incubated with Click Additive Solution for 30 min. Subsequently, the nuclei were staining with Hoechst 33342 solution. Then, the EdU-positive cells were observed by a fluorescence microscope (Olympus, Tokyo, Japan). The quantification was analyzed by Image J software (1.52a, National Institutes of Health, USA).
Migration Assay
The transwell cell culture inserts with 8 μm pores (Corning, USA) were used to conduct the cell migration assay. The cells (RSC96 and pSCs) were suspended in serum-free media and loaded into the top chamber of a 24-well transwell plate. In the bottom chamber, 20% fetal bovine serum medium was added. After 12 hours, cotton swabs were used to wipe out the non-migration cells. Later, migration cells adhered to the lower chamber of the membrane were fixed with 4% paraformaldehyde for 30 minutes, then stained with hematoxylin for 30 minutes. A microscope (Olympus, Japan) was used to count the number of cells in five random optical fields.
Enzyme-Linked Immunosorbent Assay (ELISA)
5×105 cells/well were seeded in 6-well plates. After 48 h, the cells were washed with PBS and incubated in FBS free medium. And then the supernatants were collected for ELISA. Nerve Growth Factor (NGF) and Neurotrophin 3 (NT-3) levels were determined in supernatants using an ELISA kit (SEA105Ra (NGF), SEA106Ra (NT3), Cloud-Clone Corp., China) according to the instructions of the manufacturer.
Animals
Male SD rats (200–250 g) were maintained according to US National Institutes of Health guidelines. Animal care and use procedures were approved by the Laboratory Animal Ethical and Welfare Committee of Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Ethical number: XHEC-F-2023-020). This study was conducted in compliance with the Guide for the Care and Use of Laboratory Animals published by the National Academy Press (NIH, revised in 1996). Rats with similar and good growth status were randomly divided into four groups (Sham, PBS, Nor-Exos, or Hypo-Exos-treated group) or another four groups (Hypo-Exos, Hypo+circRNA_Nkd2OE-Exos, Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos, or the NC group) in different parts of the experiment. There were 6 rats in each group.
Construction the Facial Nerve Injury (FNI) Model and Implantation of Exosomes
The rat facial nerve injury model was performed as previously published studies described.20,21 In brief, the animals were anesthetized with 30 mg/kg of 0.2% pentobarbital intraperitoneally. Crush injuries were made to the right extracranial portion of the facial nerve trunk after it was exposed, using mosquito forceps to clamp about 2 mm in length with one power grip for 60s. The loss of the blink reflex and Vibrissae Movements (VMs) were used to confirm the loss of facial nerve functions on the operated side.22 In the following stages, exosomes were implanted and the wound closed with sutures. SD rats were randomly divided into different groups (n = 6 per group). 20μL exosomes (1mg/mL) or an equal volume of PBS were injected into the proximal and distal sites of the injured nerve immediately after modeling. The facial nerve function was examined at 0, 3, 7, and 28 days after surgery. Additionally, rats were sacrificed in 28 days after surgery for modeling, and their facial nerves were collected for immunofluorescence and immunohistochemistry (IHC) staining.
Analysis of VMs
28 days after FNI, VMs recovery was measured by a digital camera (Sony, Japan). The frontal-occipital line (perpendicular to the line connecting the two orbital angles) was defined as the medial sagittal line. An angle of rostral openness was measured between the Frontal-Occipital line and the most frontal hair shaft. The maximal difference between retraction and protraction was recorded as the amplitude degree. Referring to the previously published research,23 the facial nerve function recovery was assessed by examination of VMs using the following scoring system: Record as 0 for no movement, 1 for barely detectable movement, 2 for slight movement, 3 for significant but asymmetric movement, and 4 for symmetric movement. We obtained scores of the observed VMs after FNI at 0, 3, 7, and 28 days, and compared them by statistical methods to the different treatment groups.
Immunofluorescence Analysis
Rat facial nerve segments were isolated and embedded. Cryostat sections of approximately 4 µm were then generated. Permeabilization with Triton X-100 (Beyotime, China) for 20 minutes followed by blocking with 5% goat serum for 1 hour. Next, the sections were incubated overnight at 4 °C with primary antibody, including rabbit anti‐S100β antibody (1:100, ab41548, Abcam, UK), rabbit anti-neurofilament antibody (1:100, ab8135, Abcam, UK), and rabbit anti-ki67 antibody (1:100, ab8135, Abcam, UK) followed by secondary antibody (Alexa Fluor 555 or 488-labeled Donkey Anti-Rabbit IgG (H+L), Beyotime, China) in dark area for 1.5 h at 37°C. After counterstaining with 4′,6-diamidino-2-phenylindole (DAPI, 1:1000, Thermo Scientific, US) for 15 min, tissue images were captured with fluorescence microscopy (Olympus, Tokyo, Japan).
IHC Staining
The facial nerve tissue segments were fixed in 4% paraformaldehyde overnight and embedded in paraffin, and 4 µm sagittal sections were generated with a cryostat. The sections were incubated with rabbit anti-MBP antibody (1:200, Proteintech, China) and rabbit anti-MED19 antibody (1:200, 1314-1-AP, Proteintech, China) overnight at 4 °C. The next day, secondary antibodies were added, and the samples were observed under a fluorescence microscopy (Olympus, Tokyo, Japan).
RNA Extraction, Reverse Transcription, and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Cellular RNA was extracted using the Trizol reagent (Invitrogen, USA), and exosomal RNA was extracted using the Exosome RNA Purification Kit (Simgen, China). circRNA inverse transcription was carried out using HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (R312-01, Vazyme, China). miRNA inverse transcription was carried out using a miRNA 1st Strand cDNA Synthesis Kit (MR101-01, Vazyme, China). Total RNA inverse transcription was conducted using HiScript II QRT SuperMix for qPCR (R222-01, Vazyme, China). qRT-PCR was performed using SYBRs Green PCR Master Mix (Q131-02, Vazyme, China) on Roche instruments (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the endogenous control. U6 snRNA was used as negative control for miRNA. The 2−ΔΔCt method was used as relative quantification measure of differential expression.24 The specific primers used for these analyses are listed in Supplementary Table 1.
Cell Transfection
circRNA_Nkd2 overexpression plasmid (p-circRNA) and its empty vector pcDNA3.1(+), rno-miR-214-3p mimics, inhibitor and the negative control (NC), and the lentivirus targeting circRNA_Nkd2 were bought from GenePharma (Shanghai, China). RSC96 cell lines and pSCs were seeded in six-well plates at 24 h prior to transfection with pcDNA3.1(+), p-circRNA, and rno-miR-214-3p mimics or inhibitor under 50–60% cell confluence using Lipofectamine 2000 (2,536,760, Invitrogen; ThermoFisher Scientific, Inc.) according to the guideline of the manufacturer. Gene expression was detected by qRT-PCR within 48 hours, and protein expression by Western blotting within 72 hours.
Competing Endogenous RNAs (ceRNAs) Analysis
The most significantly upregulated circRNA was identified through exosome qRT-PCR. The circRNA-miRNA-mRNA network was constructed to predict potential interactions between the selected circRNA and its targeted miRNAs based on miRanda. Cytoscape software (http://www.cytoscape.org/) was used for network import and visualization.
RNase R Treatment
The stability of circRNA was examined by RNase R treatment experiments. A total RNA extract was isolated from SCs and divided into two groups that were treated with RNase R and without RNase R at 37 °C for 30 minutes, respectively. RNase R was inactivated by incubating RNA at 70 °C for ten minutes, after which reverse transcription was performed for qRT-PCR.
RNA Fluorescence in situ Hybridization (FISH)
For facial nerve tissue, A FITC-labeled specific probe for circRNA_Nkd2 (5“-GCGTTTGGTTTGTATCGCCTTGAAG-3”, FITC) and Cy3-labeled specific probe for rno-miR-214-3p (5“-CTGCCTGTCTGTGCCTGCTGT-3”, Cy3) were designed and synthesized by Thermo Fisher Scientific. For SCs, a Cy3-labeled specific probe for circRNA_Nkd2 (Cy3, 5“-GUUUGGUUUGUAUCGCCUUGAAGCGAAAGCCUG-3”) was designed and synthesized by Sangon Biotech (Shanghai, China). A FISH experiment was performed according to the instructions of the manufacturer of FISH Kit for RNA (R0306S, Beyotime, China). After fixation and permeabilization with 0.25% Triton X-100, cells were hybridized overnight with circRNA_Nkd2 probe in hybridization buffer at 37°C. Nuclei were counterstained with DAPI. Cells were imaged using a confocal microscope (Leica, Germany).
For tissue, the fresh facial nerve tissue segments were fixed in 4% paraformaldehyde for 2 h, and the subsequent experimental steps were the same as the cell FISH.
Dual‑luciferase Reporter Assay
Sequences of wild type (WT) or mutant type (MUT) circRNA_Nkd2 or the full length of the 3′-UTR of MED19 with WT or MUT putative binding sites were interposed into the GP-miRGLO vector from GenePharma Corp (Shanghai, China). 293 T cells were seeded into 24-well plates underwent co-transfection with rno-miR-214-3p mimics or mimics NC and WT or MUT plasmids using Lipofectamine 2000 (2,536,760, Invitrogen, ThermoFisher Scientific, Inc.). Cells were harvested for luciferase detection using the Dual-Luciferase Reporter Assay Kit from GenePharma Corp and a microplate reader (Synergy HTX) according to the manufacturer’s protocol.
Western Blotting (WB)
In brief, total cellular proteins were lysed with a radio-immunoprecipitation assay (RIPA). Equal concentrations of the proteins were loaded on SDS-PAGE gel (10%). Following transfer to polyvinylidene difluoride (PVDF) membranes, samples were blocked for one hour with 5% skimmed milk solution. Next, membrane was incubated with TSG101 rabbit antibody (1:1000, ab125011, Abcam, UK), CD63 rabbit antibody (1:2000, A19023, ABclonal, China), CD81 rabbit antibody (1:1000, ab109201, Abcam, UK), MED19 rabbit antibody (1:1000, 1314-1-AP, Proteintech, China) or GAPDH rabbit antibody (1:1000, ab8245, Abcam, UK) at 4°C overnight. After washing with TBST for 4 times, the membranes were incubated with horseradish peroxidase-linked secondary biotinylated antibodies for 1.5h at room temperature. Following washing with PBST for 4 times, the membranes were then probed with HRP-labeled secondary antibodies at indoor temperature for 1 h and the images were recorded in the Gel Imaging System.
Statistics
Data are expressed as Mean ± standard deviation. One-way ANOVA or repeated measures ANOVA were used to test statistical significance of differences among groups. Student’s t-test or paired t-test was used for the comparisons between two groups. P < 0.05 was considered statistically significant.
Results
Hypoxia Enhances Exosome Release from BMSCs, and Exosomes Were Internalized by SCs
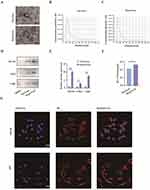
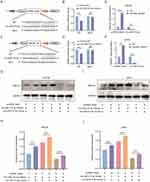
The identification of BMSCs and pSCs is displayed in Supplemental Figure 1A–J. BMSCs were incubated under hypoxic (1% O2) or normoxic conditions, respectively. Exosomes were isolated from serum-free media after 48 h of incubation, and transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and WB were used for exosome identification. TEM revealed typically rounded nanoparticles enclosed by a bilayer membrane under both Nor-Exos and Hypo-Exos (Figure 1A), with diameters ranging 50–150 nm. NTA exhibited a similar size distribution in both groups (Figure 1B and C). WB indicated that exosome surface markers CD63, CD81, and TSG101 were highly expressed in both Nor-Exos and Hypo-Exos, with the Hypo-Exos group showing higher expression levels (Figure 1D and E). Additionally, the BCA assay showed that the concentration of exosomal proteins from Hypo-Exos significantly exceeded that from the Nor-Exos groups (Figure 1F). These outcomes confirm our ability to isolate and culture purified exosomes of BMSC origin, and show that hypoxia pretreatment can promote exosome release from BMSCs without altering exosome morphology or surface markers. Furthermore, to investigate whether exosome can be taken up by SCs, Dil dye labeled exosomes were cocultured with RSC96 cells and pSCs for 24 h and monitored in real time using confocal microscopy. The results indicated that exosomes were internalized by RSC96 cells and pSCs (Figure 1G).
Hypo-Exos Enhance SCs Proliferation, Migration and Paracrine Activity
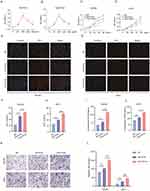
The effects of Nor-Exos and Hypo-Exos on SCs proliferation, migration, and paracrine activities were determined at the optimum concentration. Various concentrations of exosomes (50 ug/mL, 100 μg/mL, 200 μg/mL and 400 μg/mL) under normoxia and hypoxia pretreatments were cocultured with RSC96 for 48 h. Subsequently, a CCK-8 assay was conducted to determine cell viability. The results suggested that both Nor-Exos and Hypo-Exos treated cells showed optimal cell viability at exosome concentration of 100 μg/mL (Figure 2A and B). Thus, we used a concentration of 100 μg/mL for subsequent in vitro experiments. CCK-8 and EdU assays revealed that both Nor-Exos- and Hypo-Exos-treated groups exhibited superior cell proliferation compared with the negative control group (NC) in both RSC96 and pSCs. Notably, the Hypo-Exos-treated group showed more pronounced proliferation-promoting effects than Nor-Exos-treated group (Figure 2C–H). Results from the transwell cell migration assay indicated a significant increase in the number of migrating cells in both Nor-Exos and Hypo-Exos-treated groups compared with the NC group in RSC96 and pSCs, with the Hypo-Exos-treated group demonstrating a greater number of migrating cells (Figure 2K and L). ELISA revealed significantly elevated concentrations of neurotrophic factors, NGF and NT-3, in both Nor-Exos and Hypo-Exos-treated group compared with the NC group in RSC96 and pSCs, with the Hypo-Exos-treated group exhibiting higher NGF and NT-3 concentrations than the Nor-Exos-treated group (Figure 2I and J and Supplementary Figure 2A and B). These findings underscore the capacity of BMSC-derived exosomes to promote SCs proliferation, migration, and paracrine functionality, with hypoxia-primed exosomes demonstrating superior effects. Thus, Hypo-Exos may play a pivotal role in FNI repair.
Hypo-Exos Restore Facial Nerve Function Post-FNI
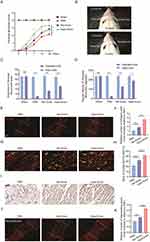
To investigate the potential of Hypo-Exos to enhance FNI repair in vivo, we conducted functional assessments of the facial nerve in four groups of rats (Sham, PBS, Nor-Exos, or Hypo-Exos-treated group). Our findings revealed a significant decrease in VMs in rats 3 days post-FNI. However, by the 4th postoperative week, rats treated with both Nor-Exos and Hypo-Exos exhibited significantly better recovery of facial nerve vibrissae activity than that in rats in the PBS group. Remarkably, FNI rats treated with Hypo-Exos displayed superior recovery compared with rats treated with Nor-Exos. Facial nerve function in the sham group remained normal with no significant changes(Figure 3A).
We further assessed the recovery of VM amplitude and angular velocity using digital photo recording. Four weeks post-surgery, significant differences in the rostral open angle between the frontal hair shaft and frontal occipital line during protraction and retraction were observed on the intact side, whereas the operated side of the PBS group remained paralyzed with only slight VMs (Figure 3B). The mean amplitude and angular velocity of the operated side in the PBS, Nor-Exos, and Hypo-Exos-treated groups were lower than those of the contralateral side. Transplantation with exosomes mitigated the damage, with the Hypo-Exos-treated groups exhibiting superior effects compared with the Nor-Exos-treated groups (Figure 3C and D). Collectively, these findings highlight the significant potential of Hypo-Exos transplantation in promoting the functional recovery of FNI.
Transplantation with Hypo-Exos Enhanced SCs Proliferation and Remyelination in vivo
To evaluate nerve regeneration, we performed histological analyses of facial nerve segments. By the 4th week post-FNI, both the Nor-Exos and Hypo-Exos transplantation groups exhibited significantly increased SCs numbers in the injured area compared with the PBS group. Notably, the Hypo-Exos-treated group displayed a significantly higher SCs count than the Nor-Exos-treated group (Figure 3E and F). Double immunohistochemistry involving S100β and ki67 on longitudinal facial nerve sections revealed a higher percentage of ki67+ SCs in both the Nor-Exos and Hypo-Exos transplantation groups. Additionally, the ratio of ki67+ SCs significantly increased in the Hypo-Exos-treated group compared with the Nor-Exos group (Figure 3G and H). IHC results showed a higher number of positive MBP cells in both the Nor-Exos and Hypo-Exos transplantation groups compared with the PBS group, with the Hypo-Exos group exhibiting even more positive MBP cells than the Nor-Exo group (Figure 3I). These findings highlight the significant remyelination effect observed in facial nerves transplanted with Hypo-Exos. Furthermore, immunofluorescence staining for neurofilaments indicated that Hypo-Exos transplantation promoted greater axon regeneration in injured facial nerves than in the Nor-Exos group (Figure 3J and K).
circRNA Expression Patterns in Nor-Exos and Hypo-Exos
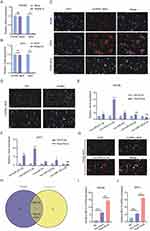
To investigate how Hypo-Exos promote FNI repair, we isolated RNA from Nor-Exos and Hypo-Exos performed microarray profiling of the circRNAs derived from the exosomes, and compared them between the two groups. Differentially expressed circRNAs from all three paired samples are shown in a volcano plot (Figure 4A), with circRNA_Nkd2 identified as the most significantly upregulated circRNA. Based on the circRNA profiling data, we selected the top five upregulated circRNAs, circRNA_Nkd2, circRNA_45863503-45,863,702-, circRNA_Pax7, circRNA_Ttc39c, and circRNA_Fam110a (Table 1), and validated their expression using qRT-PCR. As shown in Figure 4B, three circRNAs, circRNA_Nkd2, circRNA_45863503-45,863,702-, and circRNA_Ttc39c, from the five selected genes were significantly upregulated in Hypo-Exos compared with Nor-Exos. Among them, circRNA_Nkd2 was the most robustly upregulated circRNA in Hypo-Exos. Subsequently, we used Nor-Exos or Hypo-Exos to co-culture the RSC96 cell line and pSCs and detected the circRNA_Nkd2 expression levels after 48 h. qRT-PCR results showed that circRNA_Nkd2 expression was significantly upregulated in the Hypo-Exos-treated group compared with that in the Nor-Exos-treated group in both RSC96 cell line and pSCs (Figure 4C and D). These findings suggest that the exosomal circRNA_Nkd2 may play a crucial role in SCs function and facial nerve regeneration.
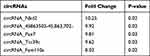 |
Table 1 Top 5 Differentially Expressed Exosomal circRNAs |
ceRNA Network Construction Based on circRNA Screening and Pathway Analysis Regulated by Exosomal circRNAs
circRNAs can act as sponges for miRNAs and indirectly regulate the expression of downstream target genes of miRNAs. The circRNA-miRNA-mRNA axis may play a key role in facial nerve regeneration-related pathways. We constructed circRNA-miRNA-mRNA networks based on five selected circRNAs, seven miRNAs, and 25 mRNAs (Figure 4E). Considering the complexity of the circRNAs and mRNAs, Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed. GO enrichment analysis results revealed that the regulation of cellular protein metabolic processes in biological processes, the luminal side of the membrane in a cellular component, and acetylcholine receptor regulator activity in molecular function were significantly enriched terms for the target genes of differentially expressed exosomal circRNAs (Figure 4G). In the KEGG pathway enrichment analysis, most exosomal circRNA target genes were enriched in the glycosaminoglycan biosynthesis-heparan sulfate/heparin signaling pathway, regulation of terpenoid backbone biosynthesis, and ribosome biogenesis in eukaryotes (Figure 4F).
Characterization and Expression Analysis of Exosomal circRNA_Nkd2
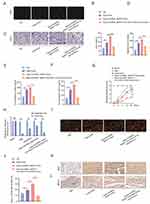
Our previous analysis focused on exosomal circRNA_Nkd2. circRNA_Nkd2 (chr1:32,066,677–32,067,055) is located on the antisense strand of Nkd2, with a length of 378 nt. First, we confirmed that circRNA_Nkd2 remained stable after incubation with RNase R in the RSC96 cell line and pSCs (Figure 5A and B). FISH staining of circRNA_Nkd2 confirmed its abundant expression in the cytoplasm of SCs (Figure 5C). Next, we examined the expression pattern of circRNA_Nkd2 in the facial nerve of the Healthy and FNI groups using FISH. We found significantly higher expression levels of circRNA_Nkd2 in the Healthy group than in the FNI group (Figure 5D), suggesting that abnormal circRNA_Nkd2 expression may be related to facial nerve repair/regeneration.
circRNA_Nkd2 Positively Regulates MED19 Expression by Sponging rno-miR-214-3p
Previous studies have shown that circRNAs in the cytoplasm competitively bind to miRNAs and regulate their target genes by acting as miRNA sponges. Therefore, we speculate that circRNA_Nkd2 targets miRNAs to modulate their downstream functions. Our microarray profiling analyzed all the predicted miRNA-binding sites and identified the top five binding miRNAs according to their binding energies (Table 2). We identified the expression levels of the top five binding miRNAs in SCs by qRT-PCR, and the results showed that rno-miR-214-3p expression was the highest in the Hypo-Exos-treated group when compared with the Nor-Exos-treated group in both the RSC96 cell line and pSCs (Figure 5E and F). The facial nerve immunofluorescence assay showed that the circRNA_Nkd2 and rno-miR-214-3p were abundantly expressed and co-localized in SCs (Figure 5G). Bioinformatics software (miRDB, https://mirdb.org/) revealed that the 3′UTR of rno-miR-214-3p contains a potential circRNA_Nkd2-binding site (Figure 6A). We performed a luciferase screening assay to verify rno-miR-214-3p binding to circRNA_Nkd2. Luciferase reporter activity was found to be significantly reduced when rno-miR-214-3p mimics were co-transfected with circRNA_Nkd2 wild-type 3′UTR sequence plasmid compared with the co-transfected mimics NC group. However, no significant difference was observed in luciferase expression when rno-miR-214-3p mimics were co-transfected with circRNA_Nkd2 mutant 3′UTR sequence plasmid compared with the co-transfected mimics NC group (Figure 6B). These results suggested that circRNA_Nkd2 functions as a rno-miR-214-3p sponge. To further confirm this, we overexpressed circRNA_Nkd2 (circRNA_Nkd2OE) in SCs and detected rno-miR-214-3p expression. The qRT-PCR results showed that rno-miR-214-3p expression was inhibited when circRNA_Nkd2 was overexpressed. These results confirmed that circRNA_Nkd2 negatively regulates rno-miR-214-3p as a sponge (Figure 6E and F).
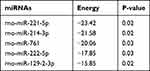 |
Table 2 Top 5 Binding miRNAs of circRNAs |
Furthermore, we used miRDB (https://mirdb.org/) and TargetScan (https://www.targetscan.org/vert_80/) to predict the downstream binding target mRNAs of rno-miR-214-3p. Three common mRNAs were screened in both the two databases of top 25: MED19, Zfand3, and Fbln5 (Figure 5H). We then performed qRT-PCR to detect Med19, Zfand3, and Fbln5 mRNA expression in RSC96 cells lines and pSCs cocultured with Nor-Exos and Hypo-Exos. Results confirmed that MED19 mRNA was highly expressed in both RSC96 cells lines and pSCs, whereas Zfand3 and Fbln5 were barely expressed. Furthermore, MED19 mRNA expression was significantly higher in the Nor-Exos and Hypo-Exos-treated groups than in the NC group, and MED19 mRNA expression in the Hypo-Exos-treated group was higher than that in the Nor-Exos-treated group (Figure 5I and J). These results indicated that MED19 was the downstream binding miRNA of rno-miR-214-3p.
To further clarify the molecular mechanisms underlying the regulation of MED19 expression by rno-miR-214-3p, we analyzed the sequence of the 3′UTR of the MED19 mRNA. Bioinformatics software (miRDB and TargetScan) revealed that the 3′UTR of MED19 contains a potential rno-miR-214-3p-binding site (Figure 6C). Luciferase reporter assay results revealed that luciferase reporter activity was significantly reduced when rno-miR-214-3p mimics were co-transfected with the MED19 wild-type plasmid compared with the co-transfected mimics NC group. Conversely, no significant difference was observed in luciferase expression when rno-miR-214-3p mimics were co-transfected with the MED19 mutant plasmid (Figure 6D). WB results showed that MED19 protein expression was upregulated by overexpression of circRNA_Nkd2 or transfection of rno-miR-214-3p inhibitor, and higher levels of MED19 protein expression were observed when circRNA_Nkd2OE plasmid and the rno-miR-214-3p inhibitor were co-transfected. MED19 protein expression was downregulated by rno-miR-214-3p mimics and rescued when circRNA_Nkd2OE plasmid and rno-miR-214-3p mimics were co-transfected (Figure 6G–J). These results confirm that circRNA_Nkd2 positively regulates MED19 expression by sponging rno-miR-214-3p.
Exosomal circRNA_Nkd2 Promoted Proliferation, Migration and Paracrine of SCs by Sponge rno-miR-214-3p in vitro
To gain deeper insights into the role of exosomal circRNA_Nkd2 in vitro, we cocultured differentially treated BMSCs-derived exosomes with SCs, including Hypo-Exos, Hypo+circRNA_Nkd2OE-Exos (BMSCs-derived exosomes overexpressing circRNA_Nkd2-Exos following hypoxic pretreatment), Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos (BMSC-derived exosomes overexpressing circRNA_Nkd2 and co-transfection with rno-miR-214-3p mimics following hypoxic pretreatment), and the NC group. In the EdU assay, we observed a higher percentage of EdU+ cells in the Hypo+circRNA_Nkd2OE-Exos-treated group than in the Hypo-Exos-treated group. However, the percentage of EdU+ cells significantly decreased in the Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos-treated group when compared with the Hypo+circRNA_Nkd2OE-Exos-treated group in both RSC96 and pSCs (Figure 7A and B and Supplementary Figure 2C and D). In the transwell cell migration assay, the cell migration numbers of the Hypo+circRNA_Nkd2OE-Exos-treated group were significantly higher than those of the Hypo-Exos-treated group. Conversely, the cell migration numbers significantly decreased in the Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos-treated group when compared with the Hypo+circRNA_Nkd2OE-Exos group (Figure 7C and D and Supplementary Figure 2E and F). Furthermore, ELISA revealed higher expression levels of neurotrophic factors (NGF and NT-3) in RSC96 and pSCs supernatants from the Hypo+circRNA_Nkd2OE-Exos-treated group when compared with the Hypo-Exos-treated group. However, the expression levels of NGF and NT-3 significantly decreased in the Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos-treated group compared with those in the Hypo+circRNA_Nkd2OE-Exos-treated group (Figure 7E and F and Supplementary Figure 2G and H). Collectively, these findings suggest that exosomal circRNA_Nkd2 promotes SCs proliferation and paracrine function by sponging rno-miR-214-3p.
Hypo-Exos Expedite SCs Proliferation, Remyelination via the circRNA_Nkd2/miR-214-3p/MED19 Axis in vivo
To gain deeper insights into the in vivo function of exosomal circRNA_Nkd2, we conducted facial nerve functional assessments in five groups of rats: Hypo-Exos, Hypo+circRNA_Nkd2OE-Exos (BMSCs-derived exosomes overexpressing circRNA_Nkd2-Exos following hypoxic pretreatment), Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos (BMSC-derived exosomes overexpressing circRNA_Nkd2 and co-transfection with rno-miR-214-3p mimics following hypoxic pretreatment), sham, and PBS groups. Results revealed that by the 4th week post-surgery, the recovery of VMs in FNI rats transplanted with Hypo-Exos and Hypo+circRNA_Nkd2OE-Exos was significantly better than that in the PBS group. Moreover, FNI rats treated with Hypo+circRNA_Nkd2OE-Exos showed significantly better recovery than the rats treated with Hypo-Exos. However, the recovery of VMs in FNI rats transplanted with Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos showed significantly poorer recovery than that of rats in the Hypo+circRNA_Nkd2OE-Exos group (Figure 7G). Similar results were observed for the amplitude of the vibrissa movement score (Figure 7H).
To evaluate nerve regeneration, we performed a histological analysis of nerve segments. After 4 weeks of FNI, compared with the PBS group, the ratio of ki67+ SCs in the facial nerves of both groups treated with Hypo-Exos and Hypo+circRNA_Nkd2OE-Exos was significantly increased, and the Hypo+circRNA_Nkd2OE-Exos-treated group showed higher ki67+ SCs than the Hypo-Exos-treated group. However, the ratio of ki67+ SCs in the facial nerves treated with Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos was significantly lower than that in the Hypo+circRNA_Nkd2OE-Exo-treated group (Figure 7I and J). IHC results showed that MBP in the Hypo+circRNA_Nkd2OE-Exos- and Hypo-Exos-treated groups was high expression compared with the PBS group. Additionally, MBP expression in the Hypo+circRNA_Nkd2OE-Exos-treated group was significantly increased compared with the Hypo-Exos-treated group, whereas transplantation of Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimic-Exos suppressed MBP expression (Figure 7K). Furthermore, IHC revealed an increased number of MED19 positive cells in both the Hypo+circRNA_Nkd2OE-Exos- and Hypo-Exos-treated groups compared with the PBS group. Notably, the Hypo+circRNA_Nkd2OE-Exos-treated group exhibited higher MED19 expression than the Hypo-Exos-treated group. Conversely, transplantation of Hypo+circRNA_Nkd2OE+rno-miR-214-3p mimics-Exos suppressed MED19 expression (Figure 7L). These results collectively demonstrate that Hypo-Exos transplantation promoted SCs proliferation, remyelination and restore facial nerve function via the circRNA_Nkd2/miR-214-3p/MED19 axis in vivo.
Discussion
Facial nerve injuries, akin to typical peripheral nerve injuries, involve shared pathological and physiological mechanisms with general peripheral nerves. SCs play a crucial role in nerve repair, which includes their influence on migration, proliferation, differentiation, and paracrine actions, ultimately dictating facial nerve function recovery25,26 Recognizing the importance of SCs involvement in neural regeneration, factors and methods that can specifically enhance SCs function may facilitate FNI recovery.27
Transplantation of mesenchymal stem cells (MSCs) has emerged as a promising strategy for addressing peripheral nerve injuries.28,29 MSCs exhibit remarkable attributes, including self-renewal, differentiation potential, and the ability to modulate hematopoietic and immune cells, rendering them attractive for tissue repair applications.6 However, direct MSC transplantation faces inherent limitations and challenges. Intravenous injection often results in the capture of most MSCs by the lungs or liver, leading to only a fraction reaching the intended target tissue.30 Moreover, concerns regarding the safety of stem cell transplantation persist, encompassing risks such as immune rejection, dedifferentiation, and tumor formation, thereby constraining the clinical utility of MSC transplantation.31 Recent studies have unveiled the therapeutic potential of extracellular vesicles, specifically exosomes secreted by stem cells, which mimic the biological functions of stem cells.32–34 Exosomes contain a variety of components, including RNA, DNA, Proteins, Amino acids, and Metabolites, primarily act as intercellular messengers, executing specific functions similar to their parent cells through paracrine function.11,35 The approach of exosome transplantation may circumvent the challenges associated with direct MSCs transplantation.36
Previous research has illuminated the therapeutic efficacy of MSC-derived exosomes in nervous system disorders such as Alzheimer’s disease,37 traumatic brain injury,38 spinal cord injury,39 and glioblastoma.40 Our experiments likewise validated the role of BMSCs-derived exosomes in promoting SCs proliferation, migration, and the secretion of nerve growth factors (NGF, and NT-3). Animal studies further underscored their contributions to remyelination, axon regeneration, and the restoration of facial nerve function.
Oxygen levels profoundly influence BMSCs proliferation, differentiation, and self-renewal. Hypoxic preconditioning of MSCs augments their biological functions and enhances therapeutic effects, given that a significant proportion of MSCs naturally inhabit hypoxic environments under natural physiological conditions.16 Previous studies have unveiled the enhanced therapeutic potential of BMSCs-derived exosomes under hypoxic conditions. For instance, hypoxia-primed BMSCs-derived exosomes were found to expedite bone formation and bolster fracture healing by transferring miR-126.17 Another study found that mesenchymal stem cell-derived exosomes mitigated cardiomyocyte apoptosis in hypoxic conditions through microRNA144, targeting the PTEN/AKT pathway.41 In our study, we demonstrated that hypoxic preconditioning of BMSCs heightens exosome secretion, with Hypo-Exos promoting SCs proliferation, migration, and paracrine function. Furthermore, we confirmed the capacity of Hypo-Exos enhanced axonal regeneration, remyelination, and the recovery of facial nerve function. Nevertheless, the precise underlying mechanisms necessitate further exploration.
A recently discovered non-coding RNA—circRNA—with a conserved loop structure formed by back-splicing, has significant relevance to various diseases and bioprocesses.42 For example, circRNA_0000253 was found to promote rat intervertebral disc degeneration by inhibiting miRNA-141-5p and downregulating SIRT1.43 In our RNA-seq analysis, several circRNAs were upregulated in Hypo-Exos. GO and KEGG enrichment analyses of these circRNAs target genes revealed associations with essential pathways such as cell proliferation, migration, paracrine, metabolism, and signal transduction, including pathways like ubiquitin-mediated proteolysis, Wnt signaling pathway, and Hippo signaling pathway, which are relevant to nerve injury repair. Among these upregulated circRNAs, circRNA_Nkd2 showed most significant increase (10.25-fold upregulation) in Hypo-Exos. Furthermore, circRNA_Nkd2 expression increased in SCs after Hypo-Exos transplantation compared with Nor-Exos, suggesting a vital role for exosomal circRNA_Nkd2 in regulating SC cellular function and repairing FNI. Our cell experiments confirmed that Hypo-Exos enhanced SCs proliferation, migration, and paracrine function, while animal experiments verified that Hypo-Exos transplantation promoted SCs proliferation, remyelination, restoration of facial nerve function after FNI.
Prior research has indicated that the molecular mechanisms of circRNAs depend on their subcellular localization, with cytoplasm circRNAs potentially acting as ceRNAs by sequestering miRNAs.44 In our study, we observed that circRNA_Nkd2 was predominantly located in the cytoplasm, suggesting that circRNA_Nkd2 may function as microRNA (miRNA) sponges, indirectly influencing downstream mRNA expression. The circRNA-miRNA-mRNA axis could play a pivotal role in facial nerve regeneration-related pathways.
Through bioinformatic analyses, we identified the circRNA_Nkd2 binding sites for rno-miR-214-3p, validated by double luciferase reporter experiments. Furthermore, we found that exosomal circRNA_Nkd2 positively regulated mediator complex subunit 19 (MED19), a target gene of rno-miR-214-3p, also confirmed using double luciferase reporter experiments. MED19, a mediator family member, has been associated with tumor development in various cancers,45,46 yet its role in nerve injury remains unexplored. Our study revealed that exosomal circRNA_Nkd2 competitively inhibits rno-miR-214-3p, resulting in increased MED19 expression. Cell experiments demonstrated that hypoxic preconditioning of BMSCs-derived circRNA_Nkd2 overexpressing exosomes enhances proliferation, migration, and paracrine function of SCs. In the FNI rat model, BMSCs-derived overexpressing exosomal circRNA_Nkd2 promoted facial nerve function recovery, but the effect was reversed by rno-miR-214-3p mimics.
Conclusion
In conclusion, we have provided evidence for a mechanism through which BMSCs-derived Hypo-Exos stimulated SCs proliferation, migration, and paracrine function to accelerate nerve regeneration via the circRNA_Nkd2/miR-214-3p/MED19 axis. Exosomal circRNA_Nkd2 upregulates MED19 expression by competitively inhibiting rno-miR-214-3p in SCs. Hypoxic pre-treatment of BMSC-derived exosomes holds promise for enhancing their therapeutic potential in FNI treatment.
Abbreviations
FNI, Facial nerve injury; SCs, Schwann cells; BMSCs, Bone mesenchymal stem cells; miRNAs, microRNAs; circRNAs, circular RNAs; MED19, Mediator complex subunit 19; pSCs, Primary Schwann cells; SD, Sprague Dawley; CCK8, Cell counting kit-8; EdU, Ethynyldeoxyuridine; OD, Optical density; IHC, Immunohistochemistry; VMs, Vibrissae Movements; qRT-PCR, Quantitative real-time polymerase chain reaction; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; FISH, Fluorescence in situ hybridization; WT, wild type; MUT, Mutant type; WB, Western Blotting; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82171360 and No. 81974186), Science and Technology Commission of Shanghai Municipality Project (No. 21Y21900500).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Engström M, Berg T, Stjernquist-Desatnik A, et al. Prednisolone and valaciclovir in Bell’s palsy: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet Neurol. 2008;7(11):993–1000. doi:10.1016/s1474-4422(08)70221-7
2. Xia W, Zhu J, Wang X, et al. ANXA1 directs Schwann cells proliferation and migration to accelerate nerve regeneration through the FPR2/AMPK pathway. FASEB J. 2020;34(10):13993–14005. doi:10.1096/fj.202000726RRR
3. Bengur FB, Stoy C, Binko MA, et al. Facial nerve repair: bioengineering approaches in preclinical models. Tissue Eng Part B Rev. 2022;28(2):364–378. doi:10.1089/ten.TEB.2020.0381
4. Boyd JG, Gordon T. Neurotrophic factors and their receptors in axonal regeneration and functional recovery after peripheral nerve injury. Mol Neurobiol. 2003;27(3):277–324. doi:10.1385/mn:27:3:277
5. Nocera G, Jacob C. Mechanisms of Schwann cell plasticity involved in peripheral nerve repair after injury. Cell Mol Life Sci. 2020;77(20):3977–3989. doi:10.1007/s00018-020-03516-9
6. Lavorato A, Raimondo S, Boido M, et al. Mesenchymal stem cell treatment perspectives in peripheral nerve regeneration: systematic review. Int J Mol Sci. 2021;22:2. doi:10.3390/ijms22020572
7. Jiang L, Jones S, Jia X. Stem cell transplantation for peripheral nerve regeneration: current options and opportunities. Int J Mol Sci. 2017;18:1. doi:10.3390/ijms18010094
8. Li X, Guan Y, Li C, et al. Immunomodulatory effects of mesenchymal stem cells in peripheral nerve injury. Stem Cell Res Ther. 2022;13(1):18. doi:10.1186/s13287-021-02690-2
9. Zhang RC, Du WQ, Zhang JY, et al. Mesenchymal stem cell treatment for peripheral nerve injury: a narrative review. Neural Regen Res. 2021;16(11):2170–2176. doi:10.4103/1673-5374.310941
10. Katsuda T, Kosaka N, Takeshita F, et al. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics. 2013;13(10–11):1637–1653. doi:10.1002/pmic.201200373
11. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:6478. doi:10.1126/science.aau6977
12. An Y, Lin S, Tan X, et al. Exosomes from adipose-derived stem cells and application to skin wound healing. Cell Prolif. 2021;54(3):e12993. doi:10.1111/cpr.12993
13. Bian D, Wu Y, Song G, et al. The application of mesenchymal stromal cells (MSCs) and their derivative exosome in skin wound healing: a comprehensive review. Stem Cell Res Ther. 2022;13(1):24. doi:10.1186/s13287-021-02697-9
14. Zhou H, Zhou J, Teng H, et al. MiR-145 enriched exosomes derived from bone marrow-derived mesenchymal stem cells protects against cerebral ischemia-reperfusion injury through downregulation of FOXO1. Biochem Biophys Res Commun. 2022;632:92–99. doi:10.1016/j.bbrc.2022.09.089
15. Chen J, Ren S, Duscher D, et al. Exosomes from human adipose-derived stem cells promote sciatic nerve regeneration via optimizing Schwann cell function. J Cell Physiol. 2019;234(12):23097–23110. doi:10.1002/jcp.28873
16. Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–161. doi:10.1016/j.stem.2010.07.007
17. Liu W, Li L, Rong Y, et al. Hypoxic mesenchymal stem cell-derived exosomes promote bone fracture healing by the transfer of miR-126. Acta Biomater. 2020;103:196–212. doi:10.1016/j.actbio.2019.12.020
18. Zhu F, Wei C, Wu H, et al. Hypoxic mesenchymal stem cell-derived exosomes alleviate ulcerative colitis injury by limiting intestinal epithelial cells reactive oxygen species accumulation and DNA damage through HIF-1α. Int Immunopharmacol. 2022;113(Pt A):109426. doi:10.1016/j.intimp.2022.109426
19. Chen Z, Tang Y, Zhu W, et al. Branched-chain aminotransferase 1 promotes Schwann cell migration and proliferation to accelerate facial nerve regeneration through the Twist/FoxC1 and Sox2 pathways. Int J Biol Macromol. 2023;242(Pt 2)):124870. doi:10.1016/j.ijbiomac.2023.124870
20. Hadlock TA, Heaton J, Cheney M, et al. Functional recovery after facial and sciatic nerve crush injury in the rat. Arch Facial Plast Surg. 2005;7(1):17–20. doi:10.1001/archfaci.7.1.17
21. Mattsson P, Janson AM, Aldskogius H, et al. Nimodipine promotes regeneration and functional recovery after intracranial facial nerve crush. J Comp Neurol. 2001;437(1):106–117. doi:10.1002/cne.1273
22. Chen JL, Li SQ, Chi FL, et al. Different sensitivities to rocuronium of the neuromuscular junctions innervated by normal/damaged facial nerves and somatic nerve in rats: the role of the presynaptic acetylcholine quantal release. Chin Med J. 2012;125(10):1747–1752.
23. Kano F, Matsubara K, Ueda M, et al. Secreted ectodomain of sialic acid-binding ig-like lectin-9 and monocyte chemoattractant protein-1 synergistically regenerate transected rat peripheral nerves by altering macrophage polarity. Stem Cells. 2017;35(3):641–653. doi:10.1002/stem.2534
24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
25. Liu B, Xin W, Tan JR, et al. Myelin sheath structure and regeneration in peripheral nerve injury repair. Proc Natl Acad Sci U S A. 2019;116(44):22347–22352. doi:10.1073/pnas.1910292116
26. Bosch-Queralt M, Fledrich R, Stassart RM. Schwann cell functions in peripheral nerve development and repair. Neurobiol Dis. 2023;176:105952. doi:10.1016/j.nbd.2022.105952
27. Xu C, Wu P, Yang K, et al. Multifunctional biodegradable conductive hydrogel regulating microenvironment for stem cell therapy enhances the nerve tissue repair. Small. 2023:e2309793. doi:10.1002/smll.202309793
28. Sharma AD, Wiederin J, Uz M, et al. Proteomic analysis of mesenchymal to Schwann cell transdifferentiation. J Proteomics. 2017;165:93–101. doi:10.1016/j.jprot.2017.06.011
29. Kizilay Z, Aktas S, Kahraman Cetin N, et al. Effect of systemic application of bone marrow-derived mesenchymal stem cells on healing of peripheral nerve injury in an experimental sciatic nerve injury model. Turk Neurosurg. 2017. doi:10.5137/1019-5149.Jtn.20811-17.1
30. Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cells. 2007;25(11):2896–2902. doi:10.1634/stemcells.2007-0637
31. Andrzejewska A, Dabrowska S, Lukomska B, et al. Mesenchymal stem cells for neurological disorders. Adv Sci (Weinh). 2021;8(7):2002944. doi:10.1002/advs.202002944
32. Lin Z, Wu Y, Xu Y, et al. Mesenchymal stem cell-derived exosomes in cancer therapy resistance: recent advances and therapeutic potential. Mol Cancer. 2022;21(1):179. doi:10.1186/s12943-022-01650-5
33. Mao Q, Liang XL, Zhang CL, et al. LncRNA KLF3-AS1 in human mesenchymal stem cell-derived exosomes ameliorates pyroptosis of cardiomyocytes and myocardial infarction through miR-138-5p/Sirt1 axis. Stem Cell Res Ther. 2019;10(1):393. doi:10.1186/s13287-019-1522-4
34. Zhu LP, Tian T, Wang JY, et al. Hypoxia-elicited mesenchymal stem cell-derived exosomes facilitates cardiac repair through miR-125b-mediated prevention of cell death in myocardial infarction. Theranostics. 2018;8(22):6163–6177. doi:10.7150/thno.28021
35. Toh WS, Lai RC, Zhang B, et al. MSC exosome works through a protein-based mechanism of action. Biochem Soc Trans. 2018;46(4):843–853. doi:10.1042/bst20180079
36. Qing L, Chen H, Tang J, et al. Exosomes and their microRNA cargo: new players in peripheral nerve regeneration. Neurorehabil Neural Repair. 2018;32(9):765–776. doi:10.1177/1545968318798955
37. Liu S, Fan M, Xu JX, et al. Exosomes derived from bone-marrow mesenchymal stem cells alleviate cognitive decline in AD-like mice by improving BDNF-related neuropathology. J Neuroinflammation. 2022;19(1):35. doi:10.1186/s12974-022-02393-2
38. Wen L, Wang YD, Shen DF, et al. Exosomes derived from bone marrow mesenchymal stem cells inhibit neuroinflammation after traumatic brain injury. Neural Regen Res. 2022;17(12):2717–2724. doi:10.4103/1673-5374.339489
39. Zhao Y, Chen Y, Wang Z, et al. Bone marrow mesenchymal stem cell exosome attenuates inflammasome-related pyroptosis via delivering circ_003564 to improve the recovery of spinal cord injury. Mol Neurobiol. 2022;59(11):6771–6789. doi:10.1007/s12035-022-03006-y
40. Wang B, Wu ZH, Lou PY, et al. Human bone marrow-derived mesenchymal stem cell-secreted exosomes overexpressing microRNA-34a ameliorate glioblastoma development via down-regulating MYCN. Cell Oncol Dordr. 2019;42(6):783–799. doi:10.1007/s13402-019-00461-z
41. Wen Z, Mai Z, Zhu X, et al. Mesenchymal stem cell-derived exosomes ameliorate cardiomyocyte apoptosis in hypoxic conditions through microRNA144 by targeting the PTEN/AKT pathway. Stem Cell Res Ther. 2020;11(1):36. doi:10.1186/s13287-020-1563-8
42. Caba L, Florea L, Gug C, et al. Circular RNA-is the circle perfect? Biomolecules. 2021;11(12). doi:10.3390/biom11121755
43. Song J, Chen ZH, Zheng CJ, et al. Exosome-transported circRNA_0000253 competitively adsorbs MicroRNA-141-5p and increases IDD. Mol Ther Nucleic Acids. 2020;21:1087–1099. doi:10.1016/j.omtn.2020.07.039
44. Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. doi:10.1186/s12943-020-01286-3
45. Weber H, Ruoff R, Garabedian MJ. MED19 alters AR occupancy and gene expression in prostate cancer cells, driving MAOA expression and growth under low androgen. PLoS Genet. 2021;17(1):e1008540. doi:10.1371/journal.pgen.1008540
46. Zhang X, Fan Y, Liu B, et al. Med19 promotes breast cancer cell proliferation by regulating CBFA2T3/HEB expression. Breast Cancer. 2017;24(3):433–441. doi:10.1007/s12282-016-0722-3
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.