Back to Journals » Cancer Management and Research » Volume 12
hsa_circRNA_012515 Is Highly Expressed in NSCLC Patients and Affects Its Prognosis
Authors Fu Y, Huang L, Tang H, Huang R
Received 10 January 2020
Accepted for publication 27 February 2020
Published 12 March 2020 Volume 2020:12 Pages 1877—1886
DOI https://doi.org/10.2147/CMAR.S245525
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Yunfeng Fu, 1,* Liang Huang, 2,* Hao Tang, 1 Rong Huang 1
1The Third Xiangya Hospital, Central South University, Changsha 410013, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hao Tang; Rong Huang
The Third Xiangya Hospital, Central South University, Changsha 410011, Hunan, People’s Republic of China
Email [email protected]; [email protected]
Background: NSCLC is one of the most common and most lethal malignancies throughout the world, and there is still a lack of sensitive diagnostic biomarkers. Identifying novel NSCLC biomarkers can help with the diagnosis and clinical decision-making.
Methods: Identifying the differentially expressed circRNAs in three cases of NSCLC tissues by high-throughput circRNA microarray sequencing. qRT-PCR was employed to detect the expression levels of hsa_circRNA_012515 in 80 cases of NSCLC tissues (tumor resection patients) and 60 cases of peripheral blood samples (chemotherapy patients), NSCLC cells and gefitinib-resistant NSCLC cell lines. Then combining with clinical data, we discussed whether it was feasible to use hsa_circRNA_012515 as the diagnostic and prognostic biomarker for NSCLC.
Results: In the cancerous tissues from NSCLC patients, NSCLC cells and gefitinib-resistant cell lines, the average expressions of hsa_circRNA_012515 increased significantly (P< 0.01). Patients of stage III–IV, with lymph node metastases, had an overexpression of hsa_circRNA_012515. High expression of hsa_circRNA_012515 was associated with lower OS and shorter PFS, and it is closely related to the prognosis of the patients. Bioinformatic analysis indicated that hsa_circRNA_012515 interacted with 5 miRNAs. This finding may shed new light on the subsequent studies on the working mechanism and functions.
Conclusion: Our study showed that hsa_circRNA_012515 may be a novel biomarker candidate for NSCLC. However, further studies are needed to ascertain the working mechanism of hsa_circRNA_012515 in the occurrence and development of NSCLC.
Keywords: non-small cell lung cancer, circRNAs, hsa_circRNA_012515, biomarker
Background
Lung cancer is one of the most common and most lethal malignancies throughout the world. Over 80% of the lung cancer patients are of non-small cell lung cancer (NSCLC), for which the 5-year survival rate is only 15%.1 In developed countries, lung cancer has become the major reason for cancer-related deaths.2 Early-stage lung cancer is usually asymptomatic.3 Therefore, 70% of the patients are already at the late stage of lung cancer or combined with local metastases upon the diagnosis.4 Early diagnosis is conducive to increasing the patients’ survival. Identifying novel biomarkers may contribute to the early diagnosis of NSCLC. Epidermal growth factor receptor (EGFR) mutations are the most common type of mutations in NSCLC,5 and its high expression is usually associated with a poor prognosis. Gefitinib is a tyrosine kinase inhibitor for EGFR (EGFR-TKI), which has been widely used in the clinical treatment of NSCLC, and its efficacy has already been recognized. However, severe resistance to EGFR-TKI has greatly restricted its clinical application.6 The resistance mechanism remains unclear for many patients.7 Identifying the targets of resistance to EGFR-TKI and clarifying the resistance mechanism will help improve the treatment effect for NSCLC patients.
Circular RNA (circRNA) is a non-coding RNA and has a more stable expression and highly conservative sequence compared with linear RNAs. CircRNAs were first found in RNA viruses.8 Along with the recent development in high-throughput sequencing and bioinformatics, it has been found that circRNAs are also expressed abundantly in eukaryotes.9 CircRNAs play an important role in the occurrence and development of many human diseases, especially cancers.10–12 circRNAs are not easily digested by exonuclease RNase and is expressed in many diseases and tissues with high stability and specificity. Thus, it is probable that circRNAs serve as a biomarker candidate.13,14 Moreover, competitive inhibition of miRNA as the molecular sponge is the most important working mechanism of circRNAs. circRNAs can absorb specific miRNA through the sponging effect, thus affecting the mRNA expression and fulfilling its biological functions.15 The above studies have shown that circRNAs potentially serve as the novel diagnostic and therapeutic biomarker candidate in cancers. In the present study, circRNA microarray sequencing was performed with qRT-PCR verification. hsa_circRNA_012515 was significantly upregulated in the gefitinib-resistant NSCLC tissues, which was in turn associated with a poor prognosis. Our results provide new clues for identifying biomarker candidates for NSCLC.
Materials and Methods
Tissue Samples
From 2015 to 2018, cancerous tissues and paracancerous tissues (>5 cm tumor margin) were collected from 83 patients with NSCLC tumor resection at our hospital. Three of these cases were selected for circRNA microarray sequencing, and then 20 and 60 cases were selected for small and large sample verification, respectively. These patients did not receive chemotherapy before surgery. At the same time, peripheral blood samples were collected from 60 patients with NSCLC during the same period after chemotherapy (gefitinib) for testing. These patients were all EGFR positive. From all patients, basic information (including age, gender, smoking history, tumor size, TNM stage, lymph node metastasis and tumor staging) was collected from the electronic medical record system. The largest diameter was determined from the CT images to calculate the tumor size. MRI and CT were, respectively, used to diagnose lymph node metastases and assess tumor differences. Tumor histological grading and staging were performed according to the WHO classification criteria and UICC grading criteria (7th edition). All tissue samples were immediately preserved at −80°C after collection.
Cell Culture
Pulmonary epithelial BEAS-2B cells and NSCLC (HCC827, H1870, H1048, PC9 and A459) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Gefitinib-resistant cell lines PC9/R and A459/R were obtained by gradually increasing the gefitinib concentration. All cells were cultured in the 1640 medium (HyClone, Logan, UT, USA) and incubated in the 5% CO2 incubator at 37°C. The log-phase cells were harvested.
RNA Extraction and Transcription
Total RNA extraction was performed from the cancerous tissues, paracancerous tissues and peripheral blood samples. Pulmonary epithelial and NSCLC cell lines using Trizol reagent (Invitrogen, USA). RNA content and quality were detected based on the absorbance at the wavelength of 260 nm and 280 nm using a spectrophotometer. Linear RNAs in the total extracted RNA were degraded using RNase R (Epicentre, USA). Amplification was applied to the residual circRNA and fluorescent cRNA was obtained by transcription.
circRNA Microarray Sequencing
Fluorescent cRNAs obtained from three pairs of tissue samples were labeled by the random labeling method with the Arraystar Super RNA Labeling Kit (Arraystar, USA). The labeled circRNA hybridized to the Arraystar Human circRNA Array V2 (8x15K, Arraystar). The microarray was scanned with Agilent Scanner G2505C. Image analysis was conducted using Agilent Feature Extraction software (version 11.0.1.1). circRNA data were analyzed using the GeneSPring 13.0 software (Agilent). Differentially expressed circRNAs were those with fold change>2.0 and P<0.05.
Verification by Quantitative qRT-PCR
Eighty pairs of tissues and 60 peripheral blood samples were processed and transcribed into cDNA. The circRNA expression was detected by real-time qRT-PCR (Arraystar) on the ViiA 7 Real-time PCR System (Applied Biosystems). In brief, a total of 10 μL random primers were added for reverse transcription of total RNA into cDNA. The reaction was initiated at 95°C (30s), followed by 40 cycles at 95°C (5 s) and 60°C (20 s). β-actin was used as an internal reference gene for qPCR. Normalization was performed against β-actin. The expression level of hsa_circRNA_012515 was calculated using the 2-ΔΔCt method (in triplicate). Primers were designed using Primer5 software for amplification of the target genes and Synthesized by Shanghai Shenggong Bioengineering Co., Ltd. The primer sequences for hsa_circRNA_012515 were 5ʹ CGTTCGAGTGTCCTGTGGAA3ʹ(F) and 5ʹ GCTTTATCATACTGCTT GCTGC3ʹ(R). The primer sequences for hsa_circRNA_092547 were 5ʹ GGCTTGTGGATCAGAATCTGAA3ʹ(F) and 5ʹ CAAAATTGGGAAAGATGAT GAA3ʹ(R). The primer sequences for hsa_circRNA_031235 were 5ʹ GGCAGAAGATCTGACAGGAT3 (F) and 5ʹGGCATCTGATGACTTTGACA3ʹ(R). The primer sequences for hsa_circRNA_068252 were 5ʹ GGTAAAGTTGCCACAGGAAG3ʹ(F) and 5ʹ GGACCATCACTTGGTGCA GT3ʹ(R). The primer sequences for hsa_circRNA_102641 were 5ʹ GTGAACTGCCTGAAGAGCTC3ʹ(F) and 5ʹ GTATCAACCCTCCTTGCT CT3ʹ(R). The primer sequences for β-actin were 5ʹ GTGGCCGAGGACTTTGATTG3ʹ(F) and 5ʹCCTGTAACAACGCATCTCATATT3ʹ(R).
Statistical Analysis
All statistical analyses were performed using SPSS 20.0 software. Data from three independent experiments were expressed as means±standard deviation. Two groups’ means were compared by t-test. ANOVA was performed to compare the means between multiple groups. Receiver operating characteristic curve (ROC) was used to determine the diagnostic value of circRNAs. When AUC was 0.5, it was considered that circRNAs had no diagnostic value. The Kaplan-Meier (K-M) survival curve was plotted and Log-rank test was used to analyze whether there was a significant difference in survival rates between patients with high and low circRNA expressions. P<0.05 indicated a significant difference.
Results
circRNA Microarray Sequencing
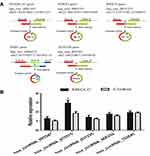
To identify specific circRNAs in NSCLS, microarray sequencing was applied to determine the circRNA expressions in three pairs of cancerous and paracancerous tissues from NSCLC patients. The results showed that 147 circRNAs were differentially expressed, including 52 upregulated circRNAs and 95 downregulated ones (Figure 1A, left insert). The differentially expressed circRNAs in the two groups were represented by the scatter plot (Figure 1B, right insert). From them, five most differentially expressed circRNAs were chosen (Table 1 and Figure 2A). Their expressions in 20 pairs of tissue samples were verified by qRT-PCR (Small sample verification). The results are shown in Figure 2B. It can be seen that hsa_circRNA_012515 was upregulated most significantly and its expression was significantly higher than that in the control group.
 |
Table 1 The Basic Information of five circRNAs Choose to Validate by qRT-PCR |
 |
Figure 1 circRNA microarray sequencing of NSCLC samples. A (left insert) is the volcano plot of circRNA expressions, and B (right insert) is the scatter plot of circRNA expressions. |
The Expression of hsa_circRNA_012515 in the NSCLC Tissues and Cell Lines Was Upregulated Significantly
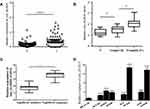
After that, the expression of hsa_circRNA_012515 was verified by qRT-PCR in 60 pairs of tissue samples (larger sample verification). It was further confirmed that the hsa_circRNA_012515 expression was significantly upregulated in NSCLC (Figure 3A) and the extent of upregulation increased with advanced tumor stage (Figure 3B). In addition, another 60 NSCLC patients received chemotherapy (gefitinib), according to the patient’s response to gefitinib treatment, 60 patients were classified as gefitinib resistance (35 cases) and gefitinib sensitivity (25 cases). We further found that the expression of hsa_circRNA_012515 in gefitinib-resistant NSCLC peripheral blood samples was significantly higher than that in gefitinib-sensitive NSCLC peripheral blood samples (Figure 3C). Furthermore, qRT-PCR verification was performed in gefitinib-resistant NSCLC cell lines, and the results were consistent with those from the clinical samples (Figure 3D). This indicated that the upregulation of hsa_circRNA_012515 may be a mechanism leading to gefitinib resistance in NSCLC patients.
The hsa_circRNA_012515 Expression Was Correlated with the Clinicopathological Features of NSCLC Patients
To better determine the clinical value of hsa_circRNA_012515, we analyzed the correlation between the expression level of hsa_circRNA_012515, clinicopathological features of NSCLC patients and the laboratory indicators. The results are shown in Table 2. It can be seen that the expression level of hsa_circRNA_012515 increased significantly in patients of stage III–IV, with lymph node metastases (P<0.05); however, the expression level was not significantly related to age, gender, smoking status, tumor size and differentiation degree of tumors. In addition, the correlation between hsa_circRNA_012515 and chemotherapy status of NSCLC patients was analyzed.
 |
Table 2 Association Between the circRNA Expression Levels and Clinicopathological Characteristics of NSCLC Surgery Patient |
hsa_circRNA_012515 Was a Good Diagnostic Biomarker and Correlated with Patients’ Prognosis
In order to determine the diagnostic value of hsa_circRNA_012515 for NSCLC patients, the ROC curve was plotted for NSCLC, as shown in Figure 4A. hsa_circRNA_012515 had a high diagnostic accuracy in NSCLC, and its AUC was 0.89 (P<0.0001). This indicated that hsa_circRNA_012515 may be a specific and sensitive diagnostic biomarker for NSCLC. Moreover, based on the average expression level of hsa_circRNA_012515, patients were divided into high and low expression groups. K-M survival analysis indicated that the overall survival (OS) (Figure 4B) and disease-free survival (DFS) (Figure 4C) were considerably shortened in patients with high expressions of hsa_circRNA_012515 than those with low expressions of hsa_circRNA_012515. This indicated that the high expression of hsa_circRNA_012515 was related to the prognosis of the patients.
miRNA Target Prediction of hsa_circRNA_012515
miRNA absorption by the sponging effect is the most important working mechanism of circRNAs. In order to determine the potential function of hsa_circRNA_012515, the self-designed miRNA target prediction software based on TargetScan and miRanda (Arraystar) was used to predict the interaction between circRNAs and miRNAs. A total of five miRNAs (hsa-miR-98-5p, hsa-miR-615-5p, hsa-let-7a-5p, hsa-let-7b-5p and hsa-let-7c-5p) were predicted to interact with hsa_circRNA_012515 (Figure 5).
 |
Figure 5 Prediction of circRNA/miRNA interaction. M: circRNA/miRNA interaction can be predicted by miRanda; T: circRNA/miRNA interaction can be predicted by TargetScan. |
Discussion
Lung cancer is the most common cancer in the clinic and also the malignancy with the highest mortality in the world.16 NSCLC is featured by high malignancy and an extremely low 5-year survival rate. This is mainly attributed to a poor understanding of the basic biology of NSCLC, which further leads to a lack of reliable biomarkers for the detection and effective drugs.17 In addition, the survival of lung cancer patients is closely related to staging. It has been shown that as the lung cancer patients evolve from stage IA to stage IV, the 5-year survival rate decreases from 82% to 6%.18 Lung cancer has a hidden onset, and its early detection is very difficult. Most of the patients are already at the late-stage upon diagnosis and have already missed the best timing for treatment. Therefore, early discovery and increasing the accuracy of early diagnosis are conducive to improving the patients’ prognosis and survival. Identifying novel NSCLC-specific biomarkers to help with the diagnosis and clinical decision-making is an urgent issue.
circRNAs are a group of endogeneous RNAs widely expressed in mammals. They are featured by stable structure and high tissue specificity, which are unlikely to be digested by exonuclease RNase.19 Therefore, circRNAs are ideal candidates for diagnostic and prognostic biomarkers for many diseases, including cancers.20 For example, Tian et al21 found that the hsa_circ_0004585 expression level was significantly increased in colorectal cancer and served as a diagnostic biomarker candidate in colorectal cancer. Zhu et al22 showed that hsa_circ_0081001 could be used as a potential biomarker and therapeutic target for osteosarcoma. Qin et al11 showed that the expression level of hsa_circ_0001649 was downregulated significantly in hepatocellular carcinoma and had a potential diagnostic value in hepatocellular carcinoma. The above studies demonstrate that circRNAs are a good diagnostic biomarker for cancers.
Early discovery and treatment are crucial for improving the prognosis of lung cancer patients.23 Better understanding of the molecular mechanism underlying the pathogenesis of NSCLC is very important for early diagnosis. Many recent studies have indicated that circRNAs play an important role in the occurrence and development of lung cancer. For example, Zhu et al24 showed that hsa_circ_0013958 could be used as a non-invasive biomarker candidate for early detection and screening of lung adenocarcinoma (a type of NSCLC). In the present study, in order to determine the expressions of circRNAs in NSCLC, we applied circRNA microarray sequencing to three pairs of cancerous and paracancerous tissues from NSCLC patients to screen for candidate circRNAs. The differentially expressed circRNAs were verified by qRT-PCR in a large amount of clinical samples, NSCLC cells and gefitinib-resistant cell lines. The results showed that the hsa_circRNA_012515 expression was significantly upregulated in the NSCLC tissues and cells, especially in the gefitinib-resistant NSCLC cells. Moreover, ROC analysis confirmed that hsa_circRNA_012515 had high specificity and sensitivity and was a good diagnostic biomarker candidate for NSCLC. It was also revealed that compared with the NSCLC patients of stage I/II, the expression level of hsa_circRNA_012515 in those of stage III/IV was upregulated significantly. In addition, the hsa_circRNA_012515 expression level was increased considerably in NSCLC patients with lymph node metastases. Thus, hsa_circRNA_012515 might be involved in the growth, progression and migration of NSCLC cells. Besides, the hsa_circRNA_012515 expression was correlated with DFS and OS, indicating that hsa_circRNA_012515 may be an important biomarker predicting poor prognosis of NSCLC patients. Taken together, hsa_circRNA_012515 has good clinical relevance, which can be applied to an early screening of NSCLC.
Regulating miRNA expressions through the sponging effect of circRNAs is an important working mechanism for circRNAs. According to the predictions by miRanda and TargetScan, we found that five miRNAs, including hsa-miR-98-5p, hsa-miR-615-5p, hsa-let-7a-5p, hsa-let-7b-5p and hsa-let-7c-5p might have interacted with hsa_circRNA_012515. These miRNAs might be crucial for the proliferation and metastasis of cancer cells. The previous study has shown that circRNA 100146 may influence the progression of NSCLC by regulating miR-361-3p and miR-615-5p.25 In addition, miR-98-5p can promote the development of pancreatic ductal adenocarcinoma by downregulating mitogen-activated protein 4 kinase 4 (MAP4K4) and inhibiting the downstream mitogen-activated protein kinase/extracellular regulated protein kinases (MAPK/ERK) signaling pathway.26 However, hsa-let-7a-5p, hsa-let-7b-5p and hsa-let-7c-5p play an important role in the occurrence and development of colorectal cancer, multiple myeloma and acute lymphoblastic leukemia.27–29 This finding provides a theoretical basis for understanding the working mechanism of hsa_circRNA_012515 in the occurrence and development of NSCLC.
To sum up, we established the diagnostic and prognostic values of hsa_circRNA_012515 in NSCLC. The average expressions of hsa_circRNA_012515 increased significantly in the NSCLC tissues, NSCLC cells and gefitinib-resistant NSCLC cell lines. Besides, the upregulation of hsa_circRNA_012515 was closely related to lymph node metastasis, tumor staging and prognosis of NSCLC patients. Therefore, hsa_circRNA_012515 is a good diagnostic and prognostic biomarker candidate for NSCLC.
Ethics and Consent Statement
This study was approved by the Ethics Committee of The Third Xiangya Hospital of Central South University and the First Affiliated Hospital of Nanchang University. All patients provided written informed consent. Informed consent was obtained from each subject in accordance with the Declaration of Helsinki.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Natural Science Foundation of Hunan Provincial China (No.2017JJ3463) and the National Natural Science Foundation of China (No. 81602801).
Disclosure
The authors report no declarations of interest.
References
1. Ferlay J, Shin HR, Bray E, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917.
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.v68.6
3. Qian F, Yang W, Chen Q, et al. Screening for early stage lung cancer and its correlation with lung nodule detection. J Thorac Dis. 2018;10:S846–S859. doi:10.21037/jtd
4. Reck M. What future opportunities may immuno-oncology provide for improving the treatment of patients with lung cancer? Ann Oncol. 2012;23:viii28–viii34. doi:10.1093/annonc/mds260
5. Shen H, Du G, Liu Z, et al. Assessment and prognostic analysis of EGFR mutations or/and HER2 overexpression in Uygur’s non-small cell lung cancer. Int J Clin Exp Med. 2015;8:22300–22309.
6. Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2015;373(16):1582.
7. Gadgeell SM, Gandhi L, Riely GJ, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer(AF-002JG): results from the dose- finding portion of a Phase 1/2 study. Lancet Oncol. 2014;15(10):1119–1128. doi:10.1016/S1470-2045(14)70362-6
8. Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi:10.1073/pnas.73.11.3852
9. Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi:10.1371/journal.pone.0030733
10. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633. doi:10.1038/bjc.2016.451
11. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi:10.3233/CBM-150552
12. Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi:10.1158/0008-5472.CAN-16-1883
13. Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi:10.1152/physrev.00041.2015
14. Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi:10.1038/onc.2017.361
15. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
16. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262
17. Tang W, Han M, Ruan B, et al. Overexpression of GOLPH3 is associated with poor survival in non-small-cell lung cancer. Am J Transl Res. 2016;8(4):1756–1762.
18. Goldstraw P, Chansky K, Crowley J, et al. International Association for the Study of Lung Cancer S, Prognostic Factors Committee AB, Participating I, International Association for the Study of Lung Cancer S, Prognostic Factors Committee Advisory B, Participating I, The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (Eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39–51. doi:10.1016/j.jtho.2015.09.009
19. Memczak S, Elefsinioti AJM, Torti F, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):331–338. doi:10.1038/nature11928
20. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2015;14:514–521. doi:10.1080/15476286.2015.1122162
21. Tian JH, Xi XH, Wang J, et al. CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res. 2019;11:5413–5423. doi:10.2147/CMAR.S199436
22. Zhu KP, Zhang CL, Hu JP, et al. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14(11):1513–1520. doi:10.7150/ijbs.27523
23. Reck M, Popat S, Reinmuth N, et al. Metastatic non-small cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25:iii27–iii39. doi:10.1093/annonc/mdu199
24. Zhu XL, Wang XY, Wei SZ, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi:10.1111/febs.14132
25. Chen LJ, Nan AR, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18:13. doi:10.1186/s12943-019-0943-0
26. Fu Y, Liu XC, Chen QY, et al. Downregulated miR-98-5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. J Exp Clin Cancer Res. 2018;37:130. doi:10.1186/s13046-018-0807-2
27. Ghanbari R, Mosakhani N, Sarhadi VK, et al. Simultaneous underexpression of let-7a-5p and let-7f-5p microRNAs in plasma and stool samples from early stage colorectal carcinoma. Cancer Biomarker. 2015;7(Suppl 1):39–48.
28. Xu H, Liu C, Zhang Y, et al. Let-7b-5p regulates proliferation and apoptosis in multiple myeloma by targeting IGF1R. Acta Biochim Biophys Sin. 2014;46(11):965–972. doi:10.1093/abbs/gmu089
29. Deniz M, Mohammadreza S. Antiproliferative effect of upregulation of hsa-let-7c-5p in human acute erythroleukemia cells. Cytotechnology. 2018;70(6):1509–1518. doi:10.1007/s10616-018-0241-5
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.