Back to Journals » The Application of Clinical Genetics » Volume 15
HLA-DQA1 and HLA-DQB1 Gene Polymorphism in Indonesian Children with Type I Diabetes Mellitus
Authors Soetjipto, Rochmah N , Faizi M, Hisbiyah Y , Endaryanto A
Received 6 November 2021
Accepted for publication 18 December 2021
Published 12 January 2022 Volume 2022:15 Pages 11—17
DOI https://doi.org/10.2147/TACG.S348115
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Martin Maurer
Soetjipto,1,2,* Nur Rochmah,2,3,* Muhammad Faizi,2,3 Yuni Hisbiyah,2,3 Anang Endaryanto2,3
1Department of Medical Biochemistry, Faculty of Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia; 2Doctoral Program of Medical Science, Faculty of Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia; 3Faculty of Medicine, Department of Child Health, Dr. Soetomo General Hospital, Universitas Airlangga, Surabaya, East Java, Indonesia
*These authors contributed equally to this work
Correspondence: Soetjipto
Doctoral Program of Medical Science, Faculty of Medicine, Universitas Airlangga, Mayjend Prof. Dr. Moestopo No. 6-8, Surabaya, East Java, 60286, Indonesia
Email [email protected]
Background: More than 40 genes influence the progression of type 1 diabetes mellitus (T1DM), including human leukocyte antigen (HLA) alleles. Different HLA genotype patterns result in diverse rates of T1DM development. HLA class II DR, DQ, and DP vary among different populations and ethnicities. Data on HLA polymorphism in T1DM in Indonesia are lacking. Therefore, this study was designed to evaluate the gene polymorphism of HLA-DQA1 and HLA-DQB1 in Indonesian children with T1DM.
Patients and Methods: In this study, 31 patients with T1DM and 31 controls were enrolled from April 2020 to April 2021. This study was conducted at Dr. Soetomo Hospital, Indonesia. We evaluated the gene polymorphism of HLA-DQA1 and HLA-DQB1 using polymerase chain reaction–restriction fragment length polymorphism. The primers used were as follows: for HLA-DQA1, DQAS34: 5ʹ-GGTGTAAACTTGTACCAG-3ʹ (forward) and DQAA261: 5ʹ-ATTGGTAGCAGCGGTAGA-3ʹ (reverse); for HLA-DQB1, DQBS43: 5ʹ-TGCTACT- TCACCAA(C/T)GGG-3ʹ (forward) and DQBA249: 5ʹ-GTAGTTGTGTCTGCA (C/T)AC-3ʹ (reverse).
Results: The most common HLA-DQA1 subtype in the T1DM group was 0101/0102 accounting for 67.6%, and 01/03 and 02/03 were found in the T1DM group only. Meanwhile, the most common HLA-DQB1 subtype in the T1DM group was 0301, accounting for 54.8%. Most subjects in this study were Javanese.
Conclusion: HLA-DQA1 0101/0102 and HLA-DQB1 0301 were commonly found in Indonesian children with T1DM.
Keywords: T1DM, HLA-DQA1, HLA-DQB1, gene polymorphism
Introduction
Type 1 diabetes mellitus (T1DM) is one of the most frequent chronic illnesses among children. The prevalence of T1DM is 5%–10% of all diabetes mellitus cases. Approximately 30 million people live with T1DM worldwide, with an estimated three-fold increase in prevalence by 2040.1 The prevalence of T1DM in Asia is surprisingly low, with 0.4–1.1 cases per 100,000 individuals per year.2 Studies have shown that T1DM has genetic susceptibility associated with gene polymorphisms or mutations. More than 40 genes influence the progression of T1DM, including human leukocyte antigen (HLA). Different HLA genotype patterns result in diverse rates of T1DM development. HLA class II DR, DQ, and DP vary among different populations and ethnicities.3 Associations between HLA genes and T1DM were first reported among Caucasians with class I molecules B8 or B15 1. For example, the high-risk HLA haplotypes in Caucasian populations, DRB1*03:01-DQB1*02:01 and DRB*04:01-DQB1*03:02, were found to be low in incidence in Japan and Southeast Asia.4 A study conducted in Southeast Asia has proven this finding, where the most prevalent HLA types were DQA1*0501 (50.7% vs 20.4%; RR = 3.97; Pc < 0.01), DQB1*0201 (48% vs 19.1%; RR = 3.86; Pc < 0.05), and DRB1*0301 (38.7 vs 6.8%; RR = 8.36; 95% Pc < 0.05).5
The pathophysiology of T1DM revolves around B cell destruction. It is thought to be related to autoreactive CD4+ helper and CD8+ cytotoxic T cells in response to at least four major antigens (ie, insulin derivatives, glutamic acid decarboxylase-65 [GAD-65], tyrosine phosphatase, and zinc transporter-8 [ZnT8]) present in pancreatic B cells. Antibodies found against these elements are good markers for T1DM.6–8 In India, a study on the prevalence of GAD antibody (GADA), ZnT8, and HLA and their diagnostic values in T1DM has found a low sensitivity of GADA and ZnT8. They concluded that for diagnosing T1DM, GADA is the most superior autoantibody. Its sensitivity is low; hence, it should be combined with HLA.9 This finding is similar to those found in Asian literature and contradicts those found in some Western studies.10–12
Studies on this particular area in Asia remain limited.9 Therefore, this study was designed to evaluate gene polymorphisms of HLA-DQA1 and HLA-DQB1 in Indonesian children with T1DM.
Patients and Methods
Study Design
This was a case–control study comparing HLA-DQA1 and HLA-DQB1 gene polymorphisms between children with T1DM and those without T1DM (control).
Subjects
In this study, 31 children with T1DM who were present for follow-up at the pediatric endocrine clinic at Dr. Soetomo General Hospital Surabaya were enrolled. For comparison, we included 31 healthy children without a history of T1DM who also attended the pediatric clinic at Dr. Soetomo General Hospital Surabaya as controls. The inclusion criteria for the subjects were as follows:
- The T1DM group included children diagnosed with T1DM according to the diagnostic criteria from American Diabetes Association (ADA) 2012, as follows: (1) glucosuria symptoms, ketonuria, and random plasma glucose of more than 200 mg/dL or (2) fasting blood glucose of more than 126 mg/dL with a plasma glucose level of more than 200 mg/dL 2 hours after oral glucose tolerance test (OGTT) or (3) a plasma glucose level of more than 200 mg/dL 2 hours after OGTT or (4) Glycated hemoglobin (HbA1c) of ≥6.5%. Meanwhile, the control group consisted of healthy children without a history of T1DM.
- Children aged 4–18 years.
- Children with low C-peptide or at least 1 antibody marker detected.
- Children whose parents consented to participate in the study.
Patients with T1DM who were admitted to the pediatric intensive care unit with severe disease were excluded. The exclusion criteria for the control group were as follows: (1) ongoing infection, (2) history of other autoimmune diseases, (3) history of allergy, and (4) history of malignancy. Sampling for both the T1DM and control groups was performed using a consecutive sampling method. Ethical approval was obtained by the Ethical Board Committee of Dr. Soetomo Hospital, Indonesia (approval no. 1889/KEPK/III/2020). All participants and their parents provided informed consent, and this study was conducted in accordance with the Declaration of Helsinki.
DNA Extraction and Genotyping
DNA was obtained from peripheral blood mononuclear cells with a standard method using the QIAmp DNA Mini Kit (Qiagen©). HLA-DQA1genotyping was performed using polymerase chain reaction (PCR) with forward primer DQAS34: 5ʹ-GGTGTAAACTTGTACCAG-3ʹ and reverse primer DQAA261: 5ʹ-ATTGGTAGCAGCGGTAGA-3ʹ, resulting in a 228bp DQA1 segment. Meanwhile, for HLA-DQB1, we used forward primer DQBS43: 5ʹ-TGCTACT- TCACCAA(C/T)GGG-3ʹ and reverse primer DQBA249: 5ʹ-GTAGTTGTGTCTGCA (C/T)AC-3ʹ, resulting in a 207bp DQB1 segment.13 For amplification, 35 cycles were performed using the following steps: denaturation at 94°C for 1 min, annealing at 60°C for 2 min, and extension using Taq DNA polymerase at 72°C for 3 min, with the final step after 35 cycles at 72°C for 3–10 min. Amplificated DNA was precipitated with ethanol and underwent acrylamide gel 3%.
Polymorphisms were evaluated using PCR–restriction fragment length polymorphism. To evaluate polymorphisms of HLA-DQA1, DdeI and RsaI restriction enzymes were used. Four alleles of DQA1 could be observed after restriction with DdeI: (1) DQA1*01 (113bp, 74bp, and 41bp fragments); (2) DQA1*02 (127bp and 98bp fragments); (3) DQA1*03 (228bp fragment); and (4) DQA1*04 (154bp and 74bp fragments). The RsaI enzyme was used to further differentiate DQA1*01 and DQA1*04 to DQA1*0101 (188bp), DQA1*0102 (40bp), DQA1*0103 (228bp), DQA1*0401 (185bp), DQA1*0501 (40bp), and DQA1*0601 (225bp).13 The visualization result of PCR-RFLP is shown in Figure 1.
Acyl, Hhal, Sau961, and MspI restriction enzymes were used to evaluate the polymorphism of HLA-DQB1. The Acyl enzyme produced DQB1*0501 (133bp, 46bp, 26bp, and 2bp) and 0502 (77bp, 56 bp, 48bp, and 26bp). The Sau961 enzyme produced 0503 allele (167bp, 39bp, and lbp) and 0401 or 0402 (151 bp, 39bp, and 17 bp). The Hhal enzyme produced 0602 or 0603 (104bp, 46bp, 29bp, 26bp, and 2bp), 0604 (133bp, 46bp, 26bp, and 2bp), and 0302 (108bp and 26bp). The Mspl restriction enzyme produced the following alleles: (1) DQB1*0301 (99bp, 72bp, and 36bp fragments); (2) DQB1*0303 (171bp and 36bp fragments); and (3) DQB1*0601 (207bp fragment). The Hhal enzyme was used to further differentiate DQB1*0401 and DQB1*0402 to DQB1*0401 (133bp, 72 bp, and 2bp) and DQB1*0402 (133 bp, 46bp, 26bp, and 2bp), respectively.13
Outcomes
The primary outcome of this study was the subtypes of HLA polymorphism in children with T1DM in Indonesia.
Statistical Analysis
All statistical analyses were performed using Statistical Package for Social Sciences, version 20.0. Descriptive analysis was conducted to explain the distribution of the subjects’ characteristics and HLA polymorphisms. Values are described using mean ± standard deviation, and nominal variables are described using numbers (percentages).
Results
Clinical Characteristics
The T1DM group consisted of 31 subjects (including 19 girls and 12 boys) with a mean age of 15 ± 3.88 years and body mass index (BMI) of 19.37 ± 4.46 kg/m2. The mean age at the onset of T1DM was 8 ± 3.72 years, and the mean T1DM duration was 7 ± 4.19 years. The baseline characteristics are presented in Table 1.
 |
Table 1 Baseline Characteristics of the Subjects in This Study |
Allele Frequencies of HLA-DQA1 in the T1DM and Control Groups
In this study, HLA-DQA1 polymorphism was evaluated using two restriction enzymes: DdeI and RsaI. DdeI produced *01, *02, *03, and *04 subtypes and their combinations. The most common subtype found in the control group was 0601, whereas the most common subtype in the T1DM group was 0101/0102. However, polymorphism between 01/03 and 02/03 was found only in the T1DM group. A complete list of HLA-DQA1 subtypes in the T1DM and control groups is presented in Table 2.
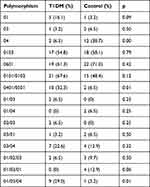 |
Table 2 Distribution of HLA-DQA1 Polymorphisms in the T1DM and Control Groups |
Allele Frequencies of HLA-DQB1 in the T1DM and Control Groups
HLA-DQB1 polymorphism was evaluated using the Acyl, Hhal, Sau961, and MspI restriction enzymes. The most common subtype found in the control group was 0502, whereas the most common subtype in the T1DM group was 0301. The subtypes 0401, 0402, 0501, 0503, 0601, and 0604 were found in both the control and T1DM groups. A complete list of HLA-DQB1 subtypes in the T1DM and control groups is shown in Table 3.
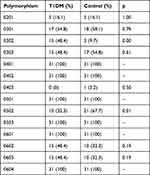 |
Table 3 Distribution HLA-DQB1 Polymorphisms in the T1DM and Control Groups |
Discussion
Most patients with T1DM in this study have HLA-DQA1 0101/0102 (67.6%); however, in the control group, most patients have HLA-DQA1 0601 (71%). Moreover, 01/04 and 01/03/01 polymorphisms were found only in the control group with a ratio of 2:4, whereas 01/03 and 02/03 was found only in the T1DM group.
The most common HLA-DQB1 polymorphism in patients with T1DM in this study was 0301 (54.8%). Furthermore, HLA-DQB1 0302, 0602, and 0603 were found in more patients with T1DM. Meanwhile, HLA-DQB1 0301, 0303, 0403, and 0502 were found in more healthy controls.
Differences in the proportion of alleles and subtypes were observed among different populations. Polymorphism HLA-DR3/DR4 is rarely found in Asia compared with that in European populations. In patients with T1DM, HLA-DQA1 0301/DQB1 0302 (HLA class II) was associated with genetic susceptibility. However, HLA-DQA1*0102/DQB1*0602 was protective for T1DM, even in individuals with HLA-DQA1*0301/DQB1*0302. Interestingly, a study by Ettinger et al has shown that HLA-DQA1*0301/DQB1*0301, HLA-DQA1*0102/DQB1*0301, and HLA-DQA1*0102/DQB1*0604 were not associated with T1DM although they have structural similarity with HLA-DQA1*0301/DQB1*0302 and HLA-DQA1*0102/DQB1*0604.14
A more recent study by Yahaya has shown that HLA-DRB1*0302–DQA1*0301 combined with DRB1*0201-DQA1*0501 increased the risk of T1DM by 20 folds, whereas the HLA-DQ6 (HLA-DQA1*0102–DQB1*0602) haplotype conferred a protective effect.1
Studies have reported that the genetic susceptibility of HLA was the most frequently reported.15–19 This genetic susceptibility varies among ethnicities.15,18,19 Studies in Iran have reported an association between HLA, gender, specificity, and age at onset. HLA-DRB1*04:01 and DQB1*03:02 alleles and DRB1*04:01–DQB1*03:02 were the most common haplotypes found in men with T1DM. Meanwhile, DRB1*03:01, DRB1*15:01, DQB1*06:01 alleles, DQB1*03:01/05:01 genotype, DRB1*03:01–DQB1*02:01, and DRB1*15:01–DQB1*06:01 were more commonly found in female with T1DM than in males.20
DRB1*03:01–DQB1*02:01 and DRB*04:01–DQB1*03:02 were lower in Japan and Southeast Asia. The most common HLA polymorphisms in Japan and Korea were DRB1*04:05–DQB1*04:01 and DRB1*09:01–DQB1*03:03.21 Meanwhile, in Arabic populations (ie, Bahrain, Lebanon, and Tunisia), the most common locus was DRB1*03:01–DQB1*02:01.22 A study by Hanscombe has shown that the combination of DRB1*07:01 and DRB1*03:03 and DQA1*03:01–DQB1*02:01 resulted in the highest risk of T1DM in African–American populations. However, in European populations, HLA-DRB1*07:01–DQA1*02:01–DQB1*02:01 was a protective factor for T1DM.3
Hamzeh has reported that 80% of patients with T1DM had HLA-DR3 or HLA-DR4 polymorphisms.23 In Japan, DR9 haplotype was the most important factor affecting the low incidence of T1DM.24–26 The profile of HLA in Indonesia is similar to the rest of South East Asian populations. The study reported that polymorphism of HLA-B*15:02 and HLA-DRB1*12:02 was frequently found in Javanese, Mollucan, and Nusa Tenggara ethnicities. Indonesia still had different characteristics of HLA compared with other South East Asian countries. Yogyakarta people had high number of DQA1*0601, DQB1*03:01, DRB1*12:02; meanwhile, Western Javanese people had HLA-A*2407. None of those HLAs are commonly found in other countries.27
Most of the patients with T1DM in this study were male, accounting for 61.3% (p = 0.45). This result contradicted Katsarou’s study that showed that T1DM is more prevalent in females than in males. Another study has reported no significant difference in T1DM prevalence between male and female patients.28
The mean age of onset in this study was relevant to the study reported by Katarou that the onset of T1DM increased with age and its peak onset was at the puberty period.29 Children with T1DM also tended to be underweight at diagnosis. Underweight is a consequence of their catabolic state, resulting in weight loss before the diagnosis.30 After receiving therapy, the weight of patients with T1DM will increase due to the anabolic effect of insulin therapy.31
The limitation of this study was having a small sample size of the participating patients and only conducted in one center. Conducting a multi-center study to obtain more data on Indonesian races is essential.
Conclusion
The most common HLA-DQA1 subtype in the T1DM group in this study was 0101/0102, whereas the most common HLA-DQB1 subtype is 0602. Further studies should be conducted to find new mutations.
Acknowledgments
The authors wish to thank the patients, family patients, and endocrine team of Dr. Soetomo Hospital, Surabaya, Indonesia. Soetjipto and Nur Rochmah are co-first authors for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yahaya T, Salisu T. Genes predisposing to type 1 diabetes mellitus and pathophysiology: a narrative review. arXiv preprint. 2020;29(1):100–109.
2. Regnell SE, Lernmark Å. Early prediction of autoimmune (type 1) diabetes. Diabetologia. 2017;60(type 1):1370–1381. doi:10.1007/s00125-017-4308-1
3. Hanscombe KB, Morris DL, Noble JA, Dilthey AT. Genetic fine mapping of Systemic Lupus Erythematosus MHC associations in Europeans and African Americans. Human Mol Genetics. 2018;(August):1–46.
4. Jahromi M. Review article Human Leukocyte Antigen (HLA) and islet autoantibodies are tools to characterize type 1 diabetes in Arab countries: emphasis on Kuwait. Disease Markers. 2019;2019(Table 1):9786078.
5. Rohana AG, Loh KC, Tin SK, et al. HLA-DQ A1, -DQB1 and -DRB1 gene polymorphism--in Malay type 1 diabetes mellitus patients and their use for riskprediction. Med J Malaysia. 2011;66(2):133-137.
6. Gabbay MAL, Sato MN, Duarte AJS, Dib SA. Serum titres of anti-glutamic acid decarboxylase- 65 and anti-IA- 2 autoantibodies are associated with different immunoregulatory milieu in newly diagnosed type 1 diabetes patients. Clin Exp Immunol. 2011;168:60–67.
7. Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383(9911):69–82. doi:10.1016/S0140-6736(13)60591-7
8. Thirunavukkarasu R, Asirvatham AJ, Chitra A. SLC30A8 gene rs13266634 C/T polymorphism in children with Type 1 diabetes in Tamil Nadu. India. 2019;11(1):55–60.
9. Singh S, Singh G, Agrawal NK, Ab Z. Prevalence of autoantibodies and HLA DR, DQ in Type 1 diabetes mellitus. J Clin Diagn Res. 2016;10:9–13.
10. Chan J, Yeung V, Chow C, et al. Pancreatic p cell function and antibodies to glutamic acid decarboxylase (anti-GAD) in Chinese patients with clinical diagnosis of insulin-dependent diabetes mellitus. Diabetes Research and Clinical Practice. 1996;32(1–2):27–34. doi:10.1016/0168-8227(96)01203-x
11. Ahmed N, Khan J, Siddiqui T. Frequency of dyslipidaemia in type 2 diabetes mellitus in patients of Hazara division frequency of dyslipidaemia in Type 2 diabetes mellitus in patients of hazara division Nasir Ahmed, Jahangir Khan, Tahir Saeed Siddiqui *. J Ayub Med Coll Abbottabad. 2014;20(2):51–54.
12. Wenzlau JM, Liu Y, Yu L, et al. A common nonsynonymous single nucleotide polymorphism in the SLC30A8 gene determines ZnT8 autoantibody specificity in Type 1 diabetes. Diabetes. 2008;57(10):2693–2697. doi:10.2337/db08-0522
13. Ikegami H, Kawaguchi Y, Yamato E, et al. Analysis by the polymerase chain reaction of histocompatibility leucocyte antigen-DRS-linked susceptibility to insulin-dependent diabetes mellitus. J Clin Endocrinol Metabol. 1992;75(November):1381–1385. doi:10.1210/jcem.75.5.1358911
14. Ettinger RA, Liu AW, Nepom GT, Kwok WW. Beta 57-Asp plays an essential role in the unique SDSstability of HLA- DQA1*0102/ DQB1*0602 alpha beta protein dimer, the class II MHC allele associatedwith protection from insulin-dependent diabetes mellitus. J Immunol. 2000;165(6):3232-3238. doi:10.4049/jimmunol.165.6.3232
15. Sperling MA, Tamborlane WV, Bettelino T, Weinzimer SA, Philip M. Diabetes mellitus. In: MA Sperling (ed.) Pediatric Endocrinology, 4th edn.Philadelphia:Elsevier Saunders; 2014:846-900.
16. Dudek NL, Purcell AW. The beta cell immunopeptidome. Beta Cell. 2014;95:115–144.
17. Van Lummel M, Van Veelen PA, De Ru AH, et al. Discovery of a selective islet peptidome presented by the highest-risk HLA-DQ8 trans Molecule. Diabetes. 2016;65:732–741.
18. Noble JA. Immunogenetics of type 1 diabetes: a comprehensive review. J Autoimmun. 2015;64:101–112. doi:10.1016/j.jaut.2015.07.014
19. Robertson CC, Rich SS. Genetics of type 1 diabetes. Curr Opin Genet Dev. 2018;50(Figure 1):7–16. doi:10.1016/j.gde.2018.01.006
20. Sayad A, Ph D, Akbari MT, Ph D, Pajouhi M. Investigation the role of gender on the HLA-DRB1 and -DQB1 association with Type 1 diabetes mellitus in Iranian patients. Cell Journal. 2013;15(2):108–115.
21. Kawabata Y, Ikegami H, Kawaguchi Y, et al. The contribution of HLA-DR and -DQ haplotypes to susceptibility to Type 1 diabetes. Diabetes. 2002;51(2):545–551. doi:10.2337/diabetes.51.2.545
22. Stayoussef M, Benmansour J, Al-jenaidi FA, et al. Influence of common and specific HLA-DRB1/DQB1 haplotypes on genetic susceptibilities of three distinct Arab populations to Type 1 diabetes mellitus. Clin Vaccine Immunol. 2009;16(1):136–138. doi:10.1128/CVI.00215-08
23. Hamzeh AR, Al Ali MT, Al-Khaja N, Al Ali MT. Association of HLA-DQA1 and -DQB1 alleles with type I diabetes in Arabs: a meta-analyses. Tissue Antigens. 2015;86(1):21–27. doi:10.1111/tan.12598
24. Jahromi MM, Eisenbarth GS. Genetic Determinants of Type 1 Diabetes. Vol. 299. IntechOpen2006; 289–299.
25. Zhang L, Gianani R, Nakayama M, Liu E, Kobayashi M. Type 1 diabetes: chronic progressive autoimmune disease. Novartis Found Symp. 2008;92:85–98.
26. Nakanishi K, Inoko H. Combination of HLA-A24, -DQA1*03, and -DR9 contributes to acute-onsetand early complete β-cell destruction in type 1 diabetes: Longitudinal study of residual β-cell function. American Diabetes Association. 2006;55(6):1862–1868. Available from: https:// diabetesjournals.org/ diabetes/article/55/6/1862/15545/Combination-of-HLA-A24-DQA1-03-and-DR9-Contributes. Accessed December 30, 2021.
27. Pradana KA, Widjaya MA, Wahjudi M. Indonesians Human Leukocyte Antigen (HLA) distributions and correlations with global diseases. Immunol Invest. 2020;49(3):333–363. doi:10.1080/08820139.2019.1673771.
28. Shojaeian A. An overview of the epidemiology of Type 1 diabetes mellitus. Int J Metab Syndr. 2020;2:1–5.
29. Katsarou A, Gudbjörnsdottir S, Rawshani A, et al. Type 1 diabetes mellitus. Nat Publ Gr. 2017;3:1–18.
30. Gregg B, Connor CG, Ruedy KJ, et al. Body mass index changes in youth in the first year after Type 1 diabetes diagnosis. J Pediatr. 2015;166(5):1265–1269.e1. doi:10.1016/j.jpeds.2015.02.036
31. De Keukelaere M, Fieuws S, Reynaert N, et al. Evolution of body mass index in children with type 1 diabetes mellitus. Eur J Ped. 2018. 177(11):1661
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

