Back to Journals » Infection and Drug Resistance » Volume 14
HIV-1 Drug Resistance and Genetic Transmission Networks Among MSM Failing Antiretroviral Therapy in South China 2014–2019
Authors Lan Y, Deng X, Li L, Cai W, Li J, Cai X, Li F, Hu F , Lei C, Tang X
Received 28 April 2021
Accepted for publication 21 July 2021
Published 4 August 2021 Volume 2021:14 Pages 2977—2989
DOI https://doi.org/10.2147/IDR.S317187
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Yun Lan,* Xizi Deng,* Linghua Li, Weiping Cai, Junbin Li, Xiaoli Cai, Feng Li, Fengyu Hu, Chunliang Lei, Xiaoping Tang
Guangzhou Eighth People’s Hospital, Guangzhou Medical University, Guangzhou, 510030, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunliang Lei; Xiaoping Tang
Guangzhou Eighth People’s Hospital, Guangzhou Medical University, 627 Dongfeng East Road, Yuexiu District, Guangzhou, 510030, People’s Republic of China
Email [email protected]; [email protected]
Background: Guangdong, located in South China, is one of the areas heavily affected by HIV-1 in China. The transmission of HIV-1 among men who have sex with men (MSM) has gradually been increasing in Guangdong.
Objective: To investigate the characteristics of the HIV-1 drug resistance, and genetic transmission networks in MSM with antiretroviral therapy (ART) failure from 2014 to 2019 in Guangdong.
Methods: HIV-1 pol gene sequences were amplified. An online subtyping tool was used to determine the genotype, and a maximum likelihood phylogenetic tree was reconstructed to confirm the genotype results. The Stanford University HIV Drug Resistance Database was used to analyse the sequences of drug resistance mutations (DRMs) and drug resistance profiles. A pairwise Tamura-Nei 93 genetic distance-based method was used to analyse the genetic transmission networks.
Results: Of 393 sequences isolated from HIV-infected MSM with ART failure, CRF01_AE (47.3%), CRF07_BC (21.4%) and CRF55_01B (21.4%) were the top three strains. 55.2% individuals harboured NRTI DRMs, whereas 67.4% carried NNRTI DRMs. 96.8% cases harboured mutations resistance to NRTIs or NNRTIs at high-level. The most common DRMs were M184I/V (42.2%), followed by V179D/E (37.9%) and K65R (27.2%). Of the subtype B sequences, no sequence fell into a cluster. Of the CRF01_AE, CRF55_01B, and CRF59_01B sequences, 14.5%, 61.9%, and 33.3% fell into clusters, respectively. Of the CRF07_BC sequences, 39.3% fell into clusters. The majority of MSM in transmission networks were concentrated at age below 35 years old, with multiple links. Moreover, approximately 54.8% of MSM had more than 2 potential transmission partners.
Conclusion: Drug resistance mutations more frequently occurred in NNRTIs among MSM with ART failure in Guangdong Province. Transmission network analysis revealed a complex transmission pattern, and more attention should be given to younger HIV-1-infected MSM with multiple links.
Keywords: HIV-1, drug resistance, genetic transmission networks, MSM
Introduction
Up to Aug 31, 2018, approximately 850 thousand people living with HIV were reported nationwide in China.1 The prevalence of HIV-1 infection is still a massive challenge to public health, although highly active antiretroviral therapy (ART) has improved the quality of life of those living with HIV or AIDS and decreased mortality rates.2 Guangdong, located in South China, is one of the areas in China that is most heavily affected by HIV-1. The number of people receiving ART has increased since the nationwide National Free Antiretroviral Treatment Program (NFATP) started in the early 2000s in Guangdong Province. HIV-infected people receiving ART have a lower risk of HIV transmission.3 However, increasing HIV drug resistance occurs after the expansion of ART and the Treat-all strategy.
Men who have sex with men (MSM) continue to be a major cause of new infections in America, Europe, Oceania, and Asia,4 including China. The proportion of newly reported HIV/AIDS cases infected via MSM increased continuously, from 0.7% in 2005 to 25.8% in 2014.5 In Guangdong Province, MSM with HIV increased from 5.0% in 2008 to 11.4% in 2013 of the infected population.6 The risk factors for HIV transmission in MSM included multiple sex partners, unprotected anal intercourse, low rates of condom use, drug use, and a history of other sexually transmitted diseases.7 More attention should be paid to MSM population, including the circulating HIV strains.
Here, we focused specifically on the occurrence and characteristics of HIV-1 drug resistance mutations (DRMs) and the extent of changes in drug susceptibility with long-term treatment among ART-experienced MSM adults in Guangdong Province, China, by targeting the polymerase (pol) gene. The results will help clarify HIV-1 genetic diversity and the progression of drug resistance among ART-failing MSM in Guangdong Province between 2014 and 2019.
Materials and Methods
Study Population and Data Collection
HIV-infected patients attending NFATP were regularly followed up at sentinel hospitals in 21 prefectures of Guangdong Province. The plasma HIV-1 viral load after 6 months of ART administration was quantified at least once per year at Guangzhou Eighth People’s Hospital. Samples with a viral load of ≥1000 copies/mL were subjected to drug resistance genotyping using an in-house RT-PCR method. Patients were eligible for inclusion in this study, including MSM living in Guangdong, followed up from Jan 1st 2014 to Dec 31st 2019, and had a plasma viral load ≥1000 copies/mL. Patients were excluded if they had used ART outside national guidelines or were missing treatment regimen information.
The epidemiological and demographic information of patients was downloaded and recorded from the National Information Surveillance System, including age, occupation, education level, marital status, WHO classification, geographical region, CD4+ T cell count, sampling area and therapeutic regimen.
Nucleic Acid Extraction, Amplification, and Sequencing
Viral RNA was extracted from 140 µL of plasma using a QIAamp Viral RNA Mini Kit (Qiagen, Germany). The amplification of the HIV-1 pol gene fragments (HXB2 2253–3318, covering the full-length protease and the first 240 amino acids of reverse transcriptase codons) was performed using one-step reverse transcription PCR (RT-PCR) by using the PrimeScript one-step RT-PCR Kit Ver. 2 (Takara, China), followed by nested PCR using Ex Taq Hot Start Version DNA Polymerase (Takara, China). The amplified PCR products were electrophoresed on a 1.0% agarose gel with the DL2000 DNA marker (Takara, China). The positive products were purified using the Agarose Gel DNA Extraction Kit (Takara, China), according to the manufacturer’s protocol and sent to Tianyi Huiyuan Genomics Company for Sanger’s sequencing.
Sequence Assembly and Phylogenetic Analyses
The obtained sequences were assembled and cleaned by DNA sequence analysis software Sequencher V5.4.6 [Gene codes, US.], and the alignments were performed by BioEdit V7.2.
To determine the genotype, the sequences were submitted to the online subtyping tool COMET developed by the Luxembourg Institute of Health (https://comet.lih.lu/).8 Then, phylogenetic analysis was conducted to confirm the genotype results.
The alignments, including all the obtained sequences, were merged with HIV-1 subtyping references retrieved from Los Alamos HIV Sequence Database using BioEdit software. Then, the maximum likelihood (ML) tree was reconstructed using PhyML software V3.09 based on the GTR substitution model. The approximate likelihood ratio test (aLRT)10 was used to estimate the branch support. The final trees were visualized using Figtree V1.4.2. The SH-like support aLTR value of 0.9 was selected as the threshold for high reliability of lineages.
Drug Resistance Analyses
The HIVdb program from Stanford University HIV Drug Resistance Database (https://hivdb.stanford.edu/hivdb/by-sequences/) was used to analyse the sequences for DRMs.11 DRMs were classified into the PI, NRTI or NNRTI drug classes. Drug resistance was divided into five levels, and sequences with a low-level or greater category of resistance were defined as drug resistance.
HIV-1 Genetic Transmission Network Construction
To avoid biases due to the potential for convergent evolution driven by antiretroviral therapy, 43 codons in PR and RT associated with DRMs were removed according to the major HIV-1 DRMs last updated on Oct 23, 2020.12
The pairwise Tamura-Nei 93 (TN93) nucleotide substitution model13 was used to calculate the genetic distance for the different genotype sequences analysed using the HyPhy software V2.2.4. The genetic transmission network was visualized and analysed using the network software Cytoscape V3.2.1 with a pairwise genetic distance of 0.005–0.015.14 The genetic distance threshold is defined as the distance that identifies the maximum number of clusters in the transmission networks.
Nucleotide Sequence Data
The nucleotide sequences were submitted to GenBank under the accession numbers MW852088-MW852480.
Statistical Analysis
An Excel database was established, and all statistical analyses were performed using IBM SPSS V25.0. Qualitative statistics were described using frequency, and quantitative statistics were described using the median (IQR). Categorical variables were compared using Fisher’s exact tests. Variables for drug resistance with a P-value < 0.05 in the Fisher’s exact tests were included in the multivariate logistic regression analysis. For all statistical tests, the level of significance for the evaluation of the two-sided P values was set at 0.05.
Results
Characteristics of the Study Population
Of all the patients who received ART for more than 6 months in Guangzhou Eighth People’s Hospital from 2014 to 2019, with once HIV-1 viral load testing per year, a total of 3895 individuals had a plasma viral load of ≥1000 copies/mL. Among the 3895 plasma samples, 3584 HIV pol gene sequences were obtained. A total of 393 sequences were from HIV-infected MSM, accounting for 11.0% (393/3584).
The demographic characteristics of these HIV-infected MSM are shown in Table 1. The participants were aged between 18 and 67 years, with a median age of 32 years, and 62.6% (246/393) of them were 15 to 35 years old. Most of them (334, 85.0%) were distributed in the Pearl River Delta region and unmarried (270, 68.7%). The median (range) baseline CD4+ T cell count was 141 (1–966) cells/mm3, and 61.8% (243/393) of the participants had a baseline CD4+ T cell count of <200 cells/mm3. The median (range) HIV-1 viral load (log10 copies/mL) in the drug resistance test was 4.37 (3.00–6.69). The dominant initial treatment regimen was Tenofovir (TDF)+ lamivudine (3TC)+ Efavirenz (EFV)/Nevirapine (NVP) (72.5%, 285/393).
 |
Table 1 Demographic Characteristics of 393 MSM-Infected Patients with Virologic Failure in ART |
Distribution of Genotypes
The ML phylogenetic analyses based on the pol gene regions showed that CRF01_AE was the most common strain circulating among MSM with ART failure in Guangdong Province (47.3%, 186/393), followed by CRF07_BC (21.4%, 84/393), CRF55_01B (21.4%, 84/393), subtype B (5.1%, 20/393), CRF55_01B (2.3%, 9/393) and others (Figure 1, Table 2).
 |
Figure 1 Distribution characteristics of HIV-1 genotypes among MSM with ART failure in Guangdong Province from 2014 to 2019. The maximum likelihood phylogenetic tree was constructed using PhyML 3.0. The reference sequences, including nine subtypes (A–D, F–H, J and K), CRF01_AE, CRF07_BC, CRF08_BC, CRF55_01B, and CRF59_01B, were downloaded from the Los Alamos HIV Sequence Database (https://www.hiv.lanl.gov/content/index). Different colour lines represent different genotypes. The subtype H sequences were selected as the outgroup. aLTR values higher than 0.9 were used to identify lineages and are indicated at the corresponding nodes of the tree. |
 |
Table 2 General Information Accompanied with Drug Resistance Among 393 MSM Patients with ART Failure |
Drug Resistance Mutations
Of the 393 MSM patients with ART failure, 69.7% (274/393) had at least one DRM. 55.2% patients (217/393) harboured NRTI resistance mutations, whereas 67.4% (265/393) carried NNRTI resistance mutations, with only 0.5% (2/393) PI resistance mutations. Dual-class mutations were present in 53.4% (210/393) of the patients, with 53.2% (209/393) having NRTI plus NNRTI resistance mutations and 0.3% (1/393) having PI plus NNRTI resistance mutations. No triple-class mutations were found.
One case (0.3%, 1/393) carried PI-related DRMs, K20T, M46I, and K43T. Among the NRTI DRMs, M184I/V (42.2%, 166/393) was the most frequent, followed by K65R (27.2%, 107/393). The most common NRTI mutation pattern was M184V+K65R, with a frequency of 15.0% (59/393). The major NNRTI DRMs were V179D/E (37.9%, 149/393), V106I/M (25.7%, 101/393), and K103N/Q (25.2%, 99/393) (Table 3).
 |
Table 3 The Number and Prevalence (%) of Drug Resistance Mutations Among MSM with ART Failure |
Resistance to ART
252 out of 393 MSM (64.1%) were resistant to at least one drug (Table 2). Among them, 244 individuals (96.8%) harboured mutations conveying high-level resistance to NRTIs or NNRTIs. 198 (78.6%) individuals expressed dual resistance to NRTIs and NNRTIs, whereas 13 (5.2%) and 33 (13.1%) ones expressed single resistance to NRTIs and NNRTIs, respectively. Fisher’s exact tests showed significant differences in drug resistance rates in terms of different treatment durations, baseline CD4+ T cell counts, and genotypes of HIV-1 (P < 0.05) (Table 2). The patients with a treatment duration of less than one year had the highest drug resistance rate (83.7%, 118/141), compared with the other group; the patients with a baseline CD4+ T cell count ≤200 cells/mm3 had a higher drug resistance rate (78.2%, 190/243) than the patients with a baseline CD4+ T cell count >200 cells/mm3; and the CRF55_01B patients had the highest drug resistance rate (81.0%, 68/84) (Table 2).
Genetic Transmission Networks of MSM with ART Failure
MSM with subtype B, CRF01_AE, CRF07_BC, CRF55_01B, and CRF59_01B pol sequences were identified and used for genetic transmission network analysis. For different genotypes, transmission networks were constructed with different genetic distance thresholds to identify the maximum number of clusters.
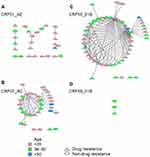
Of the 20 subtype B pol sequences, no sequence fell into a cluster at a genetic distance threshold from 0.005 to 0.015. Among 186 CRF01_AE pol sequences, 27 (14.5%) sequences fell into clusters at a genetic distance threshold of 0.015, resulting in 8 clusters ranging in size from 2 to 8 (Figure 2A). Of 84 CRF07_BC pol sequences, 33 (39.3%) sequences fell into clusters at a genetic distance threshold of 0.013, resulting in 3 clusters ranging in size from 2 to 29 (Figure 2B). Of 84 CRF55_01B pol sequences, 52 (61.9%) sequences fell into 3 clusters with sizes of 2 and 48 at a genetic distance of threshold 0.015 (Figure 2C). Of the 9 CRF59_01B pol sequences, 3 (33.3%) sequences fell into 1 cluster at a genetic distance threshold of 0.015 (Figure 2D)
Among clusters for CRF01_AE pol sequences, 17 MSM (63.0%) were found to have only one linked individual, and 10 (37.0%) had ≥2 linked individuals. Among clusters for CRF07_BC pol sequences, 16 MSM (48.5%) were found to have only one linked individual, and 17 (51.5%) had ≥2 linked individuals. Among the clusters for CRF55_01B pol sequences, 17 MSM (32.7%) were found to have only one linked individual, and 35 (67.3%) had ≥2 linked individuals. Of all 15 transmission networks, 69 (60.0%, 69/115) sequences from patients aged 15 to 35 years old were dispersed among 73.3% (11/15) of transmission networks, and 59 (51.3%, 59/115) sequences from patients with drug resistance were dispersed among 86.7% (13/15) of transmission networks (Figure 2).
There were no significant differences between genetic linkages in terms of age, occupation, marital status, WHO classification, geographical region, treatment duration, baseline CD4+ T cell count, viral load (log10) in ART failure, or individuals who showed or did not show drug resistance after ART failure (Table 4).
 |
Table 4 Distributions of HIV-1-Infected MSM with ART Failure Within the Three Major Transmission Networks |
Discussion
HIV epidemics in Guangdong have expanded beyond injection drug users and grown into sexual populations, including MSM and heterosexual contact, since 2013.15 The occurrence of HIV-1 acquired drug resistance becomes a critical clinical and public health issue and will lead to ART failure. This study analysed the prevalence of resistant variants and linking-associated factors for genetic transmission networks of MSM with ART failure in Guangdong, providing implications for the management of ART.
The results show that CRF01_AE (47.3%) and CRF07_BC (21.4%) were the dominant circulating HIV-1 strains in MSM with ART failure in Guangdong. A recent nationwide survey in China revealed that CRF01_AE and CRF07_BC accounted for more than 80% of HIV-1 infections among MSM in nine major Chinese cities,16 which was consistent with our results. CRF55_01B is the CRF01_AE/B recombinant strain originating from MSM in Shenzhen, Guangdong, and was first reported in 2013.17,18 In the present study, CRF55_01B was the third most dominant circulating genotype, accounting for 21.4%, higher than the 8.5% in Guangdong in 2013,19 9.2% in Shenzhen in 2006–2013,18 and 10.7% in Guangzhou in 2018.20
The overall prevalence of drug resistance was 66.7% in MSM with ART failure in Guangdong, which was similar to that in Yunnan (66.27% in MSM with ART failure from 2014 to 2019)21 and relatively higher than those in other areas of China, such as Sichuan (50.7% in MSM with ART failure from 2011 to 2017)22 and Hunan (41.3% in MSM with ART failure from 2012 to 2017).23
M184I/V (42.2%) was the most prevalent NRTI-associated mutation, which might be caused by the frequent use of 3TC in ART-experienced patients.24 This caused high-level resistance to 3TC and Emtricitabine (FTC) and low-level resistance to Abacavir (ABC) and Didanosine (DDI). In contrast, M184I/V increased susceptibility to Zidovudine (AZT), Stavudine (D4T) and TDF and slowed the emergence of AZT, D4T, and TDF resistance. K65R (27.2%) is the second most common NRTI-associated resistance mutation and has only a relatively low level of reduced drug susceptibility. A high frequency of K65R plus M184V/I (15.0%) was observed in this study, and K65R plus M184 V/I appeared sufficient to abrogate the NRTI activity of a regimen comprising ABC, TDF or D4T.25 As a consequence, it should be notified that more attention should be reinforced to the possibility of transmitted drug resistance, when pre-exposure prophylaxis (PrEP) was administered in this population. In our study, V179E/D was the most common NNRTI-associated mutation, accounting for 37.9%. V179E is a nonpolymorphic mutation weakly selected by NVP and EFV.26 V179D reduces NVP and EFV susceptibility by 2- to 5-fold and reduces ETR and RPV susceptibility by 2- to 3-fold, respectively.27 V179E/D does not appear to reduce the virological response to a first-line EFV-containing regimen.15 V106I/M, K103N/Q, G190A/S, and Y181C were the major NNRTI-associated mutations, similar to those in other cities in China and other countries,23,28–31 and showed broad-spectrum resistance possibly caused by the wide use of NNRTIs.
The parameters examined in this study had no significant effect on drug resistance, leading to treatment failure except treatment duration, baseline CD4+ cell count, and genotype (P<0.001, <0.001, and 0.040, respectively). Virologic failure without resistance is most often the result of inadequate drug concentrations caused by poor adherence or drug–drug interactions.32 In the present study, patients with a treatment duration below one year (83.7%) or CD4+ cell count below 200 cells/mm3 (78.2%) had a higher drug resistance rate than other groups in terms of the corresponding factors. More attention should be paid to differentiate between adherence- and resistance-driven failures, which are not mutually exclusive. In line with the evidence that different frequencies of DRMs among HIV-1 subtypes circulate among drug-naive and treated individuals in China,33 there were different drug resistance rates among different genotypes in this study (Table 2).
The genetic distance cut-off value for constructing the transmission network differs among sequences belonging to different subtypes.34 In the present study, genetic distances ≤0.013 and ≤0.015 were set as cut-off values for CRF07_BC and CRF01_AE, CRF55_01B, or CRF59_01B, respectively. The results showed that most (70.4% for CRF01_AE, 63.6% for CRF07_BC, and 55.8% for CRF55_01B) of the MSM in transmission networks were concentrated at ages below 35 years old. Moreover, approximately 54.8% (63/115) of MSM had more than 2 potential transmission partners. Individuals with multiple links could have potentially higher transmission risk.22 Therefore, more attention should be given to younger HIV-1-infected MSM with multiple links.
Conclusions
Our study focused on the genotype diversity, drug resistance and transmission networks among MSM with ART failure in Guangdong Province from 2014 to 2019 and identified CRF01_AE, CRF07_BC and CRF55_01B as the main strains circulating among MSM with ART failure. Drug resistance mutations more frequently occurred in NNRTIs. Transmission network analysis revealed a complex transmission pattern, and more attention should be paid to younger HIV-1-infected MSM with multiple links.
Ethics Approval
This study was approved by the institutional review board of the Guangzhou Eighth People’s Hospital. All participants in this study provided written informed consent.
Acknowledgments
The authors thank Prof. Xiang He from Guangdong Provincial Institute of Public Health, and Prof. Ruolei Xin from Beijing Center for Disease Prevention and Control, for their comments on this paper.
Funding
This study was supported by grants from the National Major Scientific and Technological Special Project during the 13th Five-Year Plan Period (2017ZX10202101-003), the Joint-Innovative Program in Healthcare for Special Scientific Research Projects of Guangzhou (201803040002), and Guangzhou Science and Technology Plan Project (202002030028).
Disclosure
The authors declare no conflicts of interest for this work.
References
1. NCAIDS N, China CDC. Update on the AIDS/STD epidemic in China in August, 2018. Chin J AIDS STD. 2018;24:965. doi:10.13419/j.cnki.aids.2018.10.01
2. Boyd AT, Oboho I, Paulin H, et al. Addressing advanced HIV disease and mortality in global HIV programming. AIDS Res Ther. 1999;17(1):40. doi:10.1186/s12981-020-00296-x
3. Hingankar NK, Thorat SR, Deshpande A, et al. Initial virologic response and HIV drug resistance among HIV-infected individuals initiating firstline antiretroviral therapy at 2 clinics in Chennai and Mumbai, India. Clin Infect Dis. 2012;54(suppl_4):S348–54. doi:10.1093/cid/cis005
4. Beyrer C, Abdool Karim Q. The changing epidemiology of HIV in 2013. Curr Opin HIV AIDS. 2013;8:306–310. doi:10.1097/COH.0b013e328361f53a
5. Li Z, Liao L, Feng Y, et al. Trends of HIV subtypes and phylogenetic dynamics among young men who have sex with men in China, 2009–2014. Sci Rep. 2015;5(1):16708. doi:10.1038/srep16708
6. Li Y, Lin P, Wang Y, et al. HIV/AIDS surveillance in Guangdong Province, 2000–2013. South Chin J Prev Med. 2015;41:101–106. doi:10.13217/j.scjpm.2015.0101
7. Zhang J, Guo Z, Pan X, et al. Highlighting the crucial role of Hangzhou in HIV-1 transmission among men who have sex with men in Zhejiang, China. Sci Rep. 2017;7(1):13892. doi:10.1038/s41598-017-14108-2
8. Struck D, Lawyer G, Ternes AM, et al. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014;42(18):e144. doi:10.1093/nar/gku739
9. Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi:10.1093/sysbio/syq010
10. Anisimova M, Gascuel O, Sullivan J. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi:10.1080/10635150600755453
11. Tang MW, Liu TF, Shafer RW. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology. 2012;55(2):98–101. doi:10.1159/000331998
12. Stanford University HIV drug resistance database. Major HIV-1 Drug Resistance Mutations; 2020. Available from: https://cms.hivdb.org/prod/downloads/resistance-mutation-handout/resistance-mutation-handout.pdf.
13. Wertheim JO, Kosakovsky Pond SL, Forgione LA, et al. Social and genetic networks of HIV-1 transmission in New York City. PLoS Pathog. 2017;13(1):e1006000. doi:10.1371/journal.ppat.1006000
14. Hassan AS, Pybus OG, Sanders EJ, et al. Defining HIV-1 transmission clusters based on sequence data. AIDS. 2017;31(9):1211–1222. doi:10.1097/QAD.0000000000001470
15. Yu G, Li Y, Huang X, et al. Genetic diversity and drug resistance of HIV-1 CRF55_01B in Guangdong, China. Curr HIV Res. 2020;18(3):210–218. doi:10.2174/1570162X18666200415140652
16. Han X, An M, Zhang M, et al. Identification of 3 distinct HIV-1 founding strains responsible for expanding epidemic among men who have sex with men in 9 Chinese cities. J Acquir Immune Defic Syndr. 2013;64(1):16–24. doi:10.1097/QAI.0b013e3182932210
17. Han X, An M, Zhang W, et al. Genome sequences of a novel HIV-1 circulating recombinant form, CRF55_01B, identified in China. Genome Announc. 2013;1:e00050–12. doi:10.1128/genomeA.00050-12
18. Zhao J, Cai W, Zheng C, et al. Origin and outbreak of HIV-1 CRF55_01B among MSM in Shenzhen, China. J Acquir Immune Defic Syndr. 2014;66(3):e65–7. doi:10.1097/QAI.0000000000000144
19. Zhou PP, Yu G, Kuang YQ, et al. Rapid and complicated HIV genotype expansion among high-risk groups in Guangdong Province, China. BMC Infect Dis. 2019;19(1):185. doi:10.1186/s12879-019-3788-7
20. Lan Y, He X, Li L, et al. Complicated genotypes circulating among treatment naïve HIV-1 patients in Guangzhou, China. Infect Genet Evol. 2021;87:104673. doi:10.1016/j.meegid.2020.104673
21. Ruan WQ, Liu JF, Zhang M, et al. Genotypic drug resistance of HIV-infected MSM who failed in antiviral therapy in Yunnan Province. Prev Med. 2020;3(2):
22. Yuan D, Du Z, Zhou J, et al. HIV-1 subtype diversity, drug resistance, and genetic transmission networks in men who have sex with men with virologic failure in antiretroviral therapy in Sichuan, China, 2011 to 2017. Medicine (Baltimore). 2019;98(43):e17585. doi:10.1097/MD.0000000000017585
23. Zou X, He J, Zheng J, et al. Prevalence of acquired drug resistance mutations in antiretroviral- experiencing subjects from 2012 to 2017 in Hunan Province of central South China. Virol J. 2020;17(1):38. doi:10.1186/s12985-020-01311-3
24. Vannappagari V, Ragone L, Henegar C, et al. Prevalence of pretreatment and acquired HIV-1 mutations associated with resistance to lamivudine or rilpivirine: a systematic review. Antivir Ther. 2019;24(6):393–404. doi:10.3851/IMP3331
25. Xu HT, Martinez-Cajas JL, Ntemgwa ML, et al. Effects of the K65R and K65R/M184V reverse transcriptase mutations in subtype C HIV on enzyme function and drug resistance. Retrovirology. 2009;6(1):14. doi:10.1186/1742-4690-6-14
26. Rhee SY, Gonzales MJ, Kantor R, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31(1):298–303. doi:10.1093/nar/gkg100
27. Melikian GL, Rhee SY, Varghese V, et al. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother. 2014;69(1):12–20. doi:10.1093/jac/dkt316
28. Sno R, Labadie-Bracho MY, Grünberg MG, et al. First assessment of acquired HIV-1 drug resistance and mutation patterns in suriname. AIDS Res Hum Retroviruses. 2021;37(7):557–565. doi:10.1089/AID.2020.0194
29. Omooja J, Nannyonjo M, Sanyu G, et al. Rates of HIV-1 virological suppression and patterns of acquired drug resistance among fisherfolk on first-line antiretroviral therapy in Uganda. J Antimicrob Chemother. 2019;74(10):3021–3029. doi:10.1093/jac/dkz261
30. Li T, Qian F, Yuan T, et al. Drug resistance mutation profiles of the drug-naïve and first-line regimen-treated HIV-1-infected population of Suzhou, China. Virol Sin. 2017;32(4):271–279. doi:10.1007/s12250-017-4002-y
31. Zuo L, Liu K, Liu H, et al. Trend of HIV-1 drug resistance in China: a systematic review and meta-analysis of data accumulated over 17 years (2001–2017). EClinicalMedicine. 2020;18:100238. doi:10.1016/j.eclinm.2019.100238
32. McCluskey SM, Siedner MJ, Marconi VC. Management of virologic failure and HIV drug resistance. Infect Dis Clin North Am. 2019;33(3):707–742. doi:10.1016/j.idc.2019.05.004
33. Sui H, Gui T, Jia L, et al. Different frequencies of drug resistance mutations among HIV-1 subtypes circulating in China: a Comprehensive Study. PLoS One. 2014;9(3):e91803. doi:10.1371/journal.pone.0091803
34. Zhang D, Wu J, Zhang Y, et al. Genetic characterization of HIV-1 epidemic in Anhui Province, China. Virol J. 2020;17(1):17. doi:10.1186/s12985-020-1281-y
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.