Back to Journals » Infection and Drug Resistance » Volume 17
Higher Virulence Renders K2 Klebsiella pneumoniae a Stable Share Among Those from Pyogenic Liver Abscess
Authors Dong M, Ma X , Wang D, Ma X, Zhang J, Yu L, Yang Q , Hu D , Qiao D
Received 28 September 2023
Accepted for publication 18 January 2024
Published 25 January 2024 Volume 2024:17 Pages 283—291
DOI https://doi.org/10.2147/IDR.S442454
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Min Dong,1,* Xiumin Ma,2,* Donglian Wang,3,* Xiaobo Ma,4 Jin Zhang,5 Lianhua Yu,5 Qing Yang,6 Dakang Hu,5,7 Dengyan Qiao8
1Department of Pulmonary Diseases (Department of Respiratory and Critical Care Medicine), Gansu Provincial Hospital of Traditional Chinese Medicine, Lanzhou, Gansu, People’s Republic of China; 2State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia, Clinical Laboratory Center, Tumor Hospital Affiliated to Xinjiang Medical University, Urumqi, Xinjiang, People’s Republic of China; 3Department of Laboratory Medicine, Taizhou Hospital of Zhejiang Province Affiliated to Wenzhou Medical University, Linhai, Zhejiang, People’s Republic of China; 4Department of Clinical Laboratory, the First Affiliated Hospital of Xiamen University (Xiamen Key Laboratory of Genetic Testing), Xiamen, People’s Republic of China; 5Department of Laboratory Medicine, Taizhou Municipal Hospital, Taizhou, Zhejiang, People’s Republic of China; 6Department of Laboratory Medicine, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China; 7Department of Laboratory Medicine, Huashan Hospital, Fudan University, Shanghai, People’s Republic of China; 8Department of Laboratory Medicine, Gansu Provincial Hospital of Traditional Chinese Medicine, Lanzhou, Gansu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dakang Hu; Dengyan Qiao, Email [email protected]; [email protected]
Objective: To explore why serotype K2 accounts for a stable share in Klebsiella pneumoniae from pyogenic liver abscess (PLA).
Methods: Totally 15 K2 K. pneumoniae strains from PLA, 21 K2 from non-PLA, and 31 K1 from PLA were collected from China. Sequence typing, molecular serotyping, regular PCR, and Galleria mellonella lethality were performed. A total of 12 virulence genes were detected: peg-344, allS, p-rmpA, p-rmpA2, c-rmpA, fimH, mrkD, iucA, iroN, irp2, entB, and wzi. The differences between K2 K. pneumoniae strains from PLA and non-PLA were investigated along with K1 ones.
Results: Significant differences were found between K2 strains from PLA and non-PLA for the rates of virulence genes peg-344 and iucA. The latter group also showed more diverse sequence types than the former. Significant differences were only found for virulence genes allS and irp2 between K1 and K2 strains from PLA. Based on the equal virulence factors backgrounds other than serotypes, K2 strain is more virulent than K1 as G. mellonella lethality confirmed. Gene p-rmpA only brings equal virulence to p-rmpA plus p-rmpA2 in K2 strain.
Conclusion: Based on the same virulence factors backgrounds except serotypes, K2 K. pneumoniae is more virulent than K1 from PLA, which provides a survival advantage to maintain a stable share.
Keywords: Klebsiella pneumoniae, pyogenic liver abscess, serotype, virulence, gene
Introduction
Pyogenic liver abscess (PLA) is a relatively common but fatal infectious disease, accounting for 13% of intra-abdominal infections and 48% of visceral abscesses.1–3 In China, the incidence of PLA was 17.59 per 100,000 hospital admissions, while it was 1.07 to 3.59 in the west,4 which is still on the rise.5 With the use of antibiotics and percutaneous catheter drainage, the mortality rate of PLA decreased from over 31.0% 60 years ago6 to around 10.0% in recent years.7,8 The agents of PLA include various bacteria, of which Klebsiella pneumoniae and Escherichia coli are the most documented, particularly in Asia.9,10 In the past 3 to 4 decades, K. pneumoniae has replaced E. coli as the foremost agent of PLA,11 the share of which exceeded 80.0%.12 The mean length of hospital stay for PLA was 10.3 days in New Zealand13 while it was 15.6 in China mainland.14
K. pneumoniae is now a notorious Gram-negative bacillus, causing series of infections, such as pneumonia, urinary tract infection, bacteremia, and PLA.15 As a notable member of ESKAPE, K. pneumoniae is also well known for its rapidly evolved drug resistance.16 Furthermore, K. pneumoniae is also amazing for its hypervirulence and the convergence of hypervirulence and extreme drug resistance.17 Based on the capsular polysaccharide, K. pneumoniae strains are denoted as 78 serotypes.18 For K. pneumoniae strains causing PLA, although a couple of serotypes are involved, e.g. K1, K2, K5, K16, K20, K54, K57, and K64, K1 and K2 are usually the predominant 2 types, which accounted for nearly 70.0% in sum;8,19 the ratio of K1 versus K2 is about 2:1. Therefore, K2 K. pneumoniae also plays an indispensable role in PLA. It is noted that the proportion of K1 K. pneumoniae declined, while that of K2, accounting for around 20.0%, was retained, as indicated by the 3 reports,8,12,20 which included K. pneumoniae strains from PLA during different years and regions. It is intriguing as to why K2 maintained the share over the past several years.
Typically, K1 and K2 K. pneumoniae strains are both hypervirulent.15 However, the same serotype strains from different specimen sources may differ in various backgrounds, such as virulence genes and sequence types (STs), which could also be found between different serotype strains.21 The steady-going share of K2 indicates that the traits of K2 ones from PLA need to be unveiled, in particular its virulence versus K1 ones and those of K2 from non-PLA. As Qiucheng Shi reported,22 K1 ST23 K. pneumoniae was the dominant clone in the moderate virulence-level group, although it was the most prevalent clone of the hypervirulent K. pneumoniae as Galleria mellonella lethality test was conducted; 24/36 K1 strains showed moderate virulence level, while 8/11 K2 presented high virulence level. K2 strains are generally hypervirulent with median lethal dose (LD50) <103 colony forming units (CFUs) while mouse lethality test was performed.23 However, the comparison of K1 versus K2 was not noted, although K1 strain typically shows an LD50 of 102 CFU.24 Different batches and conductors may yield different results, although mouse lethality test is done for the same strain. To unveil the traits of K2 strains from PLA, we collected 15 K2 from PLA, 21 K2 from non-PLA, and 31 K1 from PLA. Their virulence genes, factors, STs, and virulence were compared to elucidate the underlying reason for their stable share.
Materials and Methods
K. pneumoniae Strains
Totally 21 K2 K. pneumoniae strains from non-PLA were included in this study, which were collected from 5 hospitals in 4 provinces of China between January 2017 and February 2018: Huashan Hospital, 10 strains; Jinshan Hospital, 3 strains; Taizhou Municipal Hospital, 2 strains; The First Affiliated Hospital of Guangxi Medical University, 1 strain; Sixth Hospital of Shanxi Medical University, 5 strains. The other 15 K2 K. pneumoniae strains from PLA were isolated from such 3 hospitals: The First Affiliated Hospital, College of Medicine, Zhejiang University, 10 strains; The First Affiliated Hospital of Xiamen University, 3 strains; Taizhou Municipal Hospital, 2 strains. The non-PLA specimen sources included: sputum, 8; urine, 5; blood, 4; pus, 4.
The 30 K1 K. pneumoniae strains from PLA were included in this study, which were collected from 4 hospitals in 4 provinces of China between January 2017 and February 2018: Huashan Hospital, 2 strains; The First Affiliated Hospital, College of Medicine, Zhejiang University, 20 strains; The First Affiliated Hospital of Xiamen University, 8 strains. Another one was NTUH-K2044 from National Taiwan University Hospital.
Strains were identified using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system (Bruker Daltonics Inc., Fremont, CA, USA), with the standard strains ATCC 27853, ATCC 700603, and ATCC 25922 as the controls.
Strain NTUH-K2044 (GenBank accession number: AP006725.1) is a typical hypervirulent K1 K. pneumoniae, which was isolated from a patient with PLA and meningitis.25 K. pneumoniae strain HS11286 (GenBank accession number: CP003200.1), serotyped as K47, harbours blaKPC-2 and shows hypovirulence, which was isolated from a patient with pneumonia at Huashan Hospital, Fudan University, Shanghai, China.26 Strains NTUH-K2044 and HS11286 were used as controls for G. mellonella lethality tests. All the strains were kept at −80 ℃ prior to use.
Multilocus Sequence Typing (MLST)
Genomic DNA of K. pneumoniae strains were extracted using QIAamp DNA mini kit (QIAGEN Co.; Catalog number: 51,304). Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were amplified using an Applied Biosystems Veriti PCR system (ABI, San Ramon, CA, USA). The yielded products were then sequenced by an ABI 3730XL DNA Analyzer (ABI, San Ramon, CA, USA). In comparison with the K. pneumoniae MLST database website http://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=sequenceQuery, ST of each strain was obtained. The primers were shown in Table S1.
Determination of Serotypes and Virulence Genes
Serotypes K1 and K2 were identified using PCR methods.27 Virulence genes (peg-344, allS, entB, irp2, iroN, iucA, fimH, mrkD, p-rmpA2, c-rmpA, p-rmpA, and wzi)28–30 were determined using the former Veriti PCR system and agarose electrophoresis. The primers were shown in Table S1. Virulence genes p-rmpA/p-rmpA2 were used to predict the presence of virulence plasmids.
Lethality Test
Wax moth (G. mellonella) larvae lethality test was done as a reference.31 Ten larvae, weighting about 300 mg each, were injected per group with 10 μL of K. pneumoniae at the logarithmic growth phase. Surveillance time points were set at 6, 12, 24, 48, 72, and 96 h, respectively, after infection. The tested strains H2, H4, and H8 were from different lineages as pulse-field confirmed by gel electrophoresis.8
Statistical Analysis
Statistical analyses were performed using the GraphPad Prism 8 software (GraphPad Software Inc., USA). Chi-square and survival tests were used to analyze comparisons between groups; p < 0.05 was considered statistically significant.
Results
Traits of K. pneumoniae Strains Involved in G. mellonella Lethality Tests
Table 1 shows the backgrounds of the 5 K. pneumoniae strains involved in G. mellonella lethality tests, including STs, virulence genes, and predicted virulence factors. Two K2 and one K1 clinical strains were tested with NTUH-K2044 (K1) being the positive control and HS11286 (K47) being the negative control. All the 5 strains except HS11286 were from PLA. Strains H4 and H8 had same backgrounds except for serotypes.
 |
Table 1 Traits of K. pneumoniae Strains Involved in G. mellonella Lethality Tests |
Traits of K2 K. pneumoniae Strains from PLA and Non-PLA
Figure 1 shows that significant differences were only found for virulence genes peg-344 and iucA between K2 K. pneumoniae strains from PLA and non-PLA; higher rates of peg-344 and iucA were both found in K2 K. pneumoniae strains from PLA (Figure 1A). No significant differences were found in Figure 1B-D. Except for peg-344- and iucA-related virulence genes, the others and virulence plasmids were without significant differences.
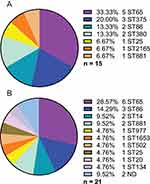
STs of K2 K. pneumoniae Strains from PLA and Non-PLA
Figure 2 shows more diverse STs in K. pneumoniae strains from non-PLA (12 STs) than those from PLA (7 STs). However, ST65 both accounted for about 30% in the two groups.
Traits of K1 and K2 K. pneumoniae Strains from PLA
Figure 3 shows that significant differences were only found for virulence genes allS and irp2 between K1 and K2 K. pneumoniae strains from PLA, which means the different rates of allantoin metabolism and yersiniabactin. The rates of allS and irp2 were both higher in K1 than in K2 strains (Figure 3A). Except for allantoin metabolism and yersiniabactin, the others and virulence plasmid were without significant differences (Figure 3B-D).
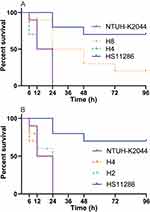
Survival Curves of G. mellonella Infected by K. pneumoniae Strains
To elucidate the virulence traits of K2 strains from PLA, K1 and K2 with rmpAs and siderophores were investigated along with the positive control NTUH-K2044 and negative control HS11286. Figure 4A suggests equal hypervirulence of NTUH-K2044 and H4, which was higher than that of H8. The lack of c-rmpA and irp2 led to less virulence of H8 than NTUH-K2044 although they were both K1. With the same backgrounds except serotypes (Table 1), more hypervirulence of H4 than that of H8 indicated K2 strains are more virulent than K1 although they are both hypervirulent.
Figure 4B confirms the equal hypervirulence of NTUH-K2044, H4, and H2. As shown in Table 1, H4 and H2 were both typed as K2 but harboured different rmpAs and siderophores. Therefore, with rmpA or rpmA2, the other rmpA/2 or excessive siderophores are redundant for the hypervirulence of K2 strains.
Discussion
The reason is still unknown as to why K2 K. pneumoniae strains maintain a stable proportion among K. pneumoniae from PLA. As revealed in this study, based on the same virulence factors backgrounds other than serotype, K2 strains are more virulent than K1 ones.
As Figure 1 shows, higher rates of peg-344 and iucA were both found in K2 K. pneumoniae strains from PLA than those from non-PLA. Gene iucA means siderophore aerobactin while the function of peg-344 remains to be elucidated.32 The less diversity of STs in K2 strains from PLA than those from non-PLA indicated the less divergent origins of the former while ST65 presented an equal share (p > 0.9999). Different from another report,23 ST66, ST373, ST374, and ST384 were absent while ST25, ST2165, and ST1881 were present in our study. As Figure 2 shows, higher rates of allS and irp2 were both found in K1 than K2 K. pneumoniae strains from PLA. Gene allS means allantoin metabolism, which facilitates the colonization of K1 strains in the gut.33 Unlike the absence of allS from another 2 reports,23,34 one K2 strain harboured it in this study. Virulence of K2 strains from PLA was rarely compared with K2 from non-PLA and K1 from PLA. According to Table 1 and Figure 4A, K2 strain H4 was more virulent than K1 strain H8. Without c-rmpA and irp2, H4 still presented the same hypervirulence with NTUH-K2044, which is likely to be the most virulent K1 strain. Figure 4B indicates that with one rmpA/2, the other rmpA/2 or excessive siderophores are redundant for the hypervirulence of K2 strains. Figure 4 suggests that based on the same virulence factors backgrounds except serotypes, K2 is more virulent than K1, which is consistent with another report.22 It should also be noted that simultaneous 3 rmpAs in K1 strains are rare.8
Since K2 K. pneumoniae strains are more virulent than K1 ones, why is that the share of K2 is less than that of K1? Although not rigorous, other related investigations may provide reasonable clues. First, the carriage of K1 may be higher. According to a report from South Korea,35 3/11 patients with PLA carried K1 K. pneumoniae strains in stool while 1/11 did K2. In healthy Korean adults, another report included 1174 Koreans and found 248 K. pneumoniae strains from stools, of which 57 were K1 (4.9%) and 54 belonged to ST23.36 Second, K1 and K2 strains always coexist with O antigen 1.37 However, different serotypes mean different capsular polysaccharides. Without the same O1 antigen, O-antigen mutant of serotype K1 became susceptible to liver clearance and caused mild abscess formation, but its serotype K2 counterpart maintained their wild-type virulence.38 These findings, together with those in this study, indicate the most important role of capsular polysaccharides in determining the higher hypervirulence of K2 strains.
This study has some limitations. First, the sample size is somewhat small due to the difficulty of collecting K2 K. pneumoniae strains from PLA. Second, virulence factors are often regulated by series of virulence genes and therefore their relations are not inevitable. Third, the same serotype strains may present diverse virulence. However, only 3 clinical strains (2 K2 and 1 K1) were included in G. mellonella lethality tests.
Taken together, based on the same virulence factors backgrounds except serotypes, K2 K. pneumoniae is more virulent than K1 one from PLA, which provides a survival advantage to maintain a stable share.
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Ethics Approval and Consent to Participate
The studies involving human participants were reviewed and approved by the Ethics Committee of Huashan Hospital (Shanghai, China) (ethical approval No. 2021-484). The consent for participation was waived for this study in accordance with the national legislation and the institutional requirements. As per Item No. 34 of Measures for Ethical Review of Biomedical Research Involving Human Beings (National Health and Family Planning Commission of China, 2014), the strains involved in this study belong to anonymous recorded materials and therefore the ethical approval was needed while consent for participation was waived. All the methods were carried out in accordance with relevant guidelines and regulations for the present study.
Acknowledgments
We thank such researchers for providing Klebsiella pneumoniae strains involved in this study: Professor Jin-Town Wang (Department of Internal Medicine, National Taiwan University Hospital), Professor Xiaofei Jiang (Department of Laboratory Medicine, Huashan Hospital, Fudan University), Gang Li (Department of Laboratory Medicine, Jinshan Hospital of Fudan University, Shanghai, China), Zehua Yang (Department of Laboratory Medicine, Sixth Hospital of Shanxi Medical University, Taiyuan, Shanxi, China), and Meng Li (Department of Clinical Laboratory, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, China).
Author Contributions
All authors made a significant contribution to the work reported, whether that was in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The work was supported by the Natural Science Foundation of Gansu (21JR11RA209) and Zhejiang Provincial Health Commission under Grant Numbers 2023KY1326 and 2024KY535.
Disclosure
The authors report there are no competing interests to declare.
References
1. Chan DS, Archuleta S, Llorin RM, Lye DC, Fisher D. Standardized outpatient management of Klebsiella pneumoniae liver abscesses. Int J Infect Dis. 2013;17(3):e185–8. doi:10.1016/j.ijid.2012.10.002
2. Rahimian J, Wilson T, Oram V, Holzman RS. Pyogenic liver abscess: recent trends in etiology and mortality. Clin Infect Dis. 2004;39(11):1654–1659. doi:10.1086/425616
3. Chen YC, Lin CH, Chang SN, Shi ZY. Epidemiology and clinical outcome of pyogenic liver abscess: an analysis from the National Health Insurance Research Database of Taiwan, 2000-2011. J Microbiol Immunol Infect. 2016;49(5):646–653. doi:10.1016/j.jmii.2014.08.028
4. Xu S, Shi BQ, Chao LM, Tan YS, Zhang XJ. Prognostic nomogram for the combination therapy of percutaneous catheter drainage and antibiotics in pyogenic liver abscess patients. Abdom Radiol. 2020;45(2):393–402. doi:10.1007/s00261-019-02359-8
5. Yin D, Ji C, Zhang S, et al. Clinical characteristics and management of 1572 patients with pyogenic liver abscess: a 12-year retrospective study. Liver Int. 2021;41(4):810–818. doi:10.1111/liv.14760
6. Huang CJ, Pitt HA, Lipsett PA, et al. Pyogenic hepatic abscess. Changing trends over 42 years. Ann Surg. 1996;223(5):600–607. doi:10.1097/00000658-199605000-00016
7. Lee SS, Chen YS, Tsai HC, et al. Predictors of septic metastatic infection and mortality among patients with Klebsiella pneumoniae liver abscess. Clin Infect Dis. 2008;47(5):642–650. doi:10.1086/590932
8. Chen H, Fang L, Chen W, et al. Pyogenic liver abscess-caused Klebsiella pneumoniae in a tertiary hospital in China in 2017: implication of hypervirulent carbapenem-resistant strains. BMC Infect Dis. 2022;22(1):685. doi:10.1186/s12879-022-07648-0
9. Wang H, Ren Y, Chang Z, Liu Z. The increased recurrence rate of liver abscess caused by extended-spectrum beta-lactamase-producing Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2020;39(7):1315–1320. doi:10.1007/s10096-020-03848-1
10. Lee JH, Jang YR, Ahn SJ, Choi SJ, Kim HS. A retrospective study of pyogenic liver abscess caused primarily by Klebsiella pneumoniae vs. non-Klebsiella pneumoniae: CT and clinical differentiation. Abdom Radiol. 2020;45(9):2669–2679. doi:10.1007/s00261-019-02389-2
11. Pope JV, Teich DL, Clardy P, McGillicuddy DC. Klebsiella pneumoniae liver abscess: an emerging problem in North America. J Emerg Med. 2011;41(5):e103–5. doi:10.1016/j.jemermed.2008.04.041
12. Qu TT, Zhou JC, Jiang Y, et al. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis. 2015;15:161. doi:10.1186/s12879-015-0899-7
13. Kubovy J, Karim S, Ding S. Pyogenic liver abscess: incidence, causality, management and clinical outcomes in a New Zealand cohort. N Z Med J. 2019;132(1492):30–35.
14. Zhang J, Du Z, Bi J, et al. Comparison of clinical characteristics and outcomes of pyogenic liver abscess patients < 65 years of age versus >/= 65 years of age. BMC Infect Dis. 2019;19(1):233. doi:10.1186/s12879-019-3837-2
15. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
16. De Oliveira DMP, Forde BM, Kidd TJ, et al. Antimicrobial Resistance in ESKAPE Pathogens. Clin Microbiol Rev. 2020;33(3). doi:10.1128/CMR.00181-19
17. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3). doi:10.1128/CMR.00001-19
18. Pan YJ, Fang HC, Yang HC, et al. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J Clin Microbiol. 2008;46(7):2231–2240. doi:10.1128/JCM.01716-07
19. Zhang S, Zhang X, Wu Q, et al. Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob Resist Infect Control. 2019;8:166. doi:10.1186/s13756-019-0615-2
20. Yeh KM, Kurup A, Siu LK, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. 2007;45(2):466–471. doi:10.1128/JCM.01150-06
21. Liao CH, Huang YT, Lai CC, et al. Klebsiella pneumoniae bacteremia and capsular serotypes, Taiwan. Emerg Infect Dis. 2011;17(6):1113–1115. doi:10.3201/eid/1706.100811
22. Shi Q, Lan P, Huang D, et al. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18(1):94. doi:10.1186/s12866-018-1236-2
23. Lin JC, Koh TH, Lee N, et al. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 2014;6:21. doi:10.1186/1757-4749-6-21
24. Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–560. doi:10.1016/j.jinf.2015.07.010
25. Wu KM, Li LH, Yan JJ, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191(14):4492–4501. doi:10.1128/JB.00315-09
26. Liu P, Li P, Jiang X, et al. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol. 2012;194(7):1841–1842. doi:10.1128/JB.00043-12
27. Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284–293. doi:10.1086/519262
28. Russo TA, Olson R, Fang CT, et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. 2018;56(9). doi:10.1128/JCM.00776-18
29. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
30. Compain F, Babosan A, Brisse S, et al. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52(12):4377–4380. doi:10.1128/JCM.02316-14
31. Tian D, Liu X, Chen W, et al. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg Microbes Infect. 2022;11(1):1936–1949. doi:10.1080/22221751.2022.2103454
32. Bulger J, MacDonald U, Olson R, Beanan J, Russo TA. Metabolite Transporter PEG344 Is Required for Full Virulence of Hypervirulent Klebsiella pneumoniae Strain hvKP1 after Pulmonary but Not Subcutaneous Challenge. Infect Immun. 2017;85(10). doi:10.1128/IAI.00093-17
33. Chou HC, Lee CZ, Ma LC, Fang CT, Chang SC, Wang JT. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun. 2004;72(7):3783–3792. doi:10.1128/IAI.72.7.3783-3792.2004
34. Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62(1):1–6. doi:10.1016/j.diagmicrobio.2008.04.007
35. Kim JH, Jeong Y, Lee CK, et al. Characteristics of Klebsiella pneumoniae Isolates from Stool Samples of Patients with Liver Abscess Caused by Hypervirulent K. pneumoniae. J Korean Med Sci. 2020;35(2):e18. doi:10.3346/jkms.2020.35.e18
36. Chung DR, Lee H, Park MH, et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis. 2012;31(4):481–486. doi:10.1007/s10096-011-1334-7
37. Hansen DS, Mestre F, Alberti S, et al. Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J Clin Microbiol. 1999;37(1):56–62. doi:10.1128/JCM.37.1.56-62.1999
38. Yeh KM, Chiu SK, Lin CL, et al. Surface antigens contribute differently to the pathophysiological features in serotype K1 and K2 Klebsiella pneumoniae strains isolated from liver abscesses. Gut Pathog. 2016;8:4. doi:10.1186/s13099-016-0085-5
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.