Back to Journals » Journal of Asthma and Allergy » Volume 16
High Transcriptional Activity and Clinical Correlations in Eosinophils of Patients with Late-Onset Asthma
Authors Lin TY , Lo CY, Chang PJ, Lo YL, Lee CS, Chang CH, Yu CT, Yao JH, Lin SM
Received 20 April 2023
Accepted for publication 14 August 2023
Published 21 August 2023 Volume 2023:16 Pages 863—878
DOI https://doi.org/10.2147/JAA.S417974
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Amrita Dosanjh
Ting-Yu Lin,1,2 Chun-Yu Lo,1,2 Po-Jui Chang,1,2 Yu-Lun Lo,1,2 Chung-Shu Lee,1– 3 Chih-Hao Chang,1– 3 Chih-Teng Yu,1,2 Jonathan Huai Yao,1 Shu-Min Lin1,2
1Department of Thoracic Medicine, Chang Gung Memorial Hospital, Taipei, Taiwan; 2College of Medicine, Chang Gung University, Taoyuan, Taiwan; 3Department of Pulmonary and Critical Care Medicine, New Taipei Municipal Tucheng Hospital, New Taipei City, Taiwan
Correspondence: Shu-Min Lin, Department of Thoracic Medicine, Chang Gung Memorial Hospital, 199 Tun-Hwa N. Road, Taipei, Taiwan, Fax + 886 33272474, Email [email protected]
Background: The immunological features of eosinophils in early-onset asthma (EOA) differ from those in late-onset asthma (LOA). Clinical trials of anti-interleukin-5 (IL-5) treatment for severe eosinophilic asthma showed a better response for LOA patients than EOA patients. We wonder if the transcriptional activity of activated eosinophils was different in EOA and LOA.
Methods: Eosinophils obtained from well-controlled EOA and LOA patients and normal subjects were compared in terms of the mRNA expression of activation-related genes and specific markers related to cell functions in eosinophils activated by IL-5 or IL-17. The correlation between mRNA expression and clinical features and lung function was further analyzed.
Results: The transcriptional expression of most genes was higher in activated eosinophils from LOA patients than in those from EOA patients and normal subjects. After IL-17 stimulation, the expression of certain genes was higher in atopic EOA patients than in non-atopic EOA patients. Similar observation was noted in obese EOA patients. After IL-5 stimulation, the transcriptional expression of most genes in eosinophils from LOA patients was negatively correlated with indicators of lung function. These correlations were less pronounced in EOA patients: After IL-17 stimulation, some genes in EOA patients were negatively correlated with post-bronchodilator changes in lung function.
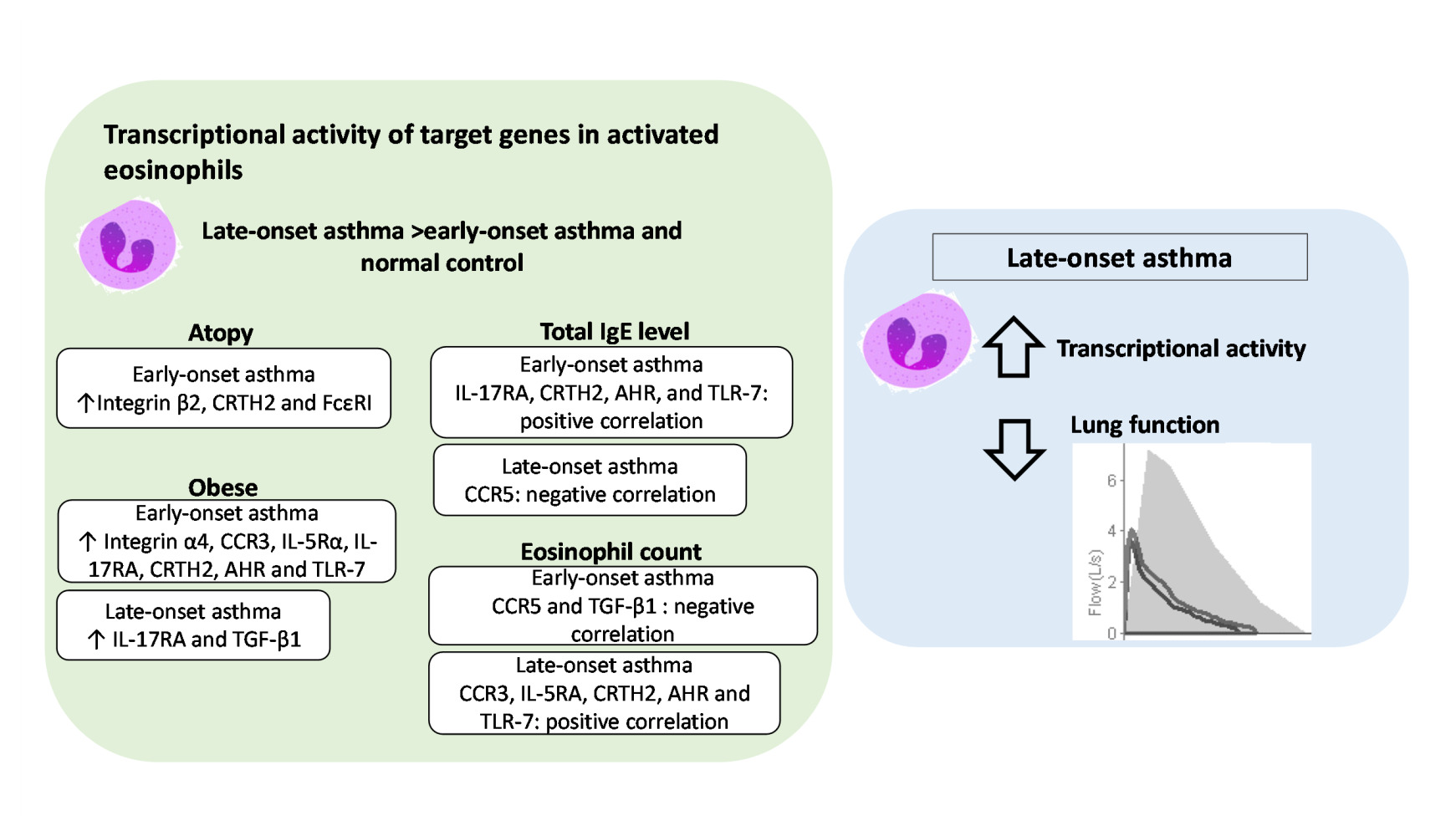
Conclusion: This study describes differences in the transcriptional active patterns of eosinophils and their correlation to atopy and obesity by age of onset. High transcriptional activity in activated eosinophils and a negative correlation to lung function indicate the importance of eosinophils in the pathogenesis of LOA.
Keywords: late-onset asthma, early-onset asthma, eosinophils, messenger RNA
Graphical Abstract:
Introduction
Airway eosinophilia is associated with poor symptom control and airway remodeling in asthma.1–3 Recruitment of eosinophils requires eosinophil activation, leading to arrest on the endothelium and extravasation, followed by recruitment to the airway. Surface proteins (eg, integrins, cytokines, and chemokine receptors) have been used to indicate the activation of circulating eosinophils.4–6 In addition to the expression of activated markers, the priming of eosinophil circulation in asthmatic patients under allergen challenge or cytokine stimulation has also been shown to induce a functional response stronger than that of the eosinophils of control subjects.7,8 Researchers have also identified correlations between the surface proteins of circulating eosinophils and the features of asthma; however, the results have been inconsistent. The possible explanation is the extravasation of activated eosinophils from circulation. It has also been suggested that in vitro experimental methods alter protein expression.6
Early-onset asthma (EOA) and late-onset asthma (LOA) are recognized as distinct phenotypes, in terms of risk factors, remission rate, co-morbidities, eosinophilic inflammation, and gene signatures.9–12 One study reported that airway eosinophils in patients with LOA were less strongly associated with type 2 inflammation than were eosinophils in patients with EOA.11 In cases of LOA, blood eosinophilia levels were also associated with disease severity.13 Interestingly, recent clinical trials involving anti-interleukin-5 (IL-5) treatment for severe eosinophilic asthma showed a better response for patients with LOA (onset after the age of 40) than for with patients with EOA.14–16 These findings suggest that there is a fundamental mechanistic difference between eosinophils in the cases of EOA and LOA. This prompted us to consider whether there were differences in the priming response of circulating eosinophils between LOA and EOA.
In order to prove this concept, we analyzed the transcriptional levels of target genes associated with activation or specific cell functions in the eosinophils from EOA and LOA following activation using IL-5 or IL-17. Finally, we examined correlations between the transcriptional levels and patient characteristics, including atopy, obesity, T2 biomarkers and lung function to identify patient-related factors associated with the transcriptomes of eosinophils.
Material and Methods
Study Design and Population
This study recruited adult asthma patients at an outpatient clinic of a medical center. Diagnosis of asthma was based on variable respiratory symptoms, such as wheeze, shortness of breath and cough, and a history of one or more of the following: 1. >12% and >200mL improvement in forced expiratory volume in 1 second (FEV1) following the use of a bronchodilator; 2. PC20 of methacholine test <8 mg/mL; and 3. Diurnal variation of >10% in peak expiratory flow. Asthma control levels were defined in accordance with guidelines outlined in the 2018 Global Initiative for Asthma (GINA).17 Pulmonary function, blood eosinophil count and immunoglobulin E (IgE) (ImmunoCAP, Phadia, Sweden) were recorded. Patients with any positive specific IgE to allergens (>0.35 KU/L) were considered atopic. Subjects were non-smokers and clinically free from respiratory or systemic infection during the two months prior to recruitment. Patients who were ≥40 years old at the time of asthma onset without a childhood history of dyspnea and frequent bouts of bronchitis were defined as LOA. Otherwise, they were considered EOA. Subjects who had normal pulmonary function, no respiratory symptoms and no history of chronic respiratory diseases were recruited as healthy control.
Ethical Statement
All patients provided written informed consent. This study was approved by the Chang Gung Medical Foundation Institutional Review Board (No. 201900943A3, approval date: October 1, 2019).
Sample Collection
Fifty milliliters of peripheral blood sampled from subjects was processed by Ficoll-Hypaque density centrifugation (GE Healthcare, Bio-science AB, Uppsala, Sweden). The lower layer consisted mainly of granulocytes and erythrocytes and was treated using an erythrocyte lysis solution (Bioman Scientific CO., LTD, New Taipei City, Taiwan). Eosinophils were isolated using an eosinophil isolation kit through the depletion of CD16+ neutrophils (Miltenyi Biotec Inc. Auburn, CA, USA) in accordance with the manufacturer’s specifications. The purification of eosinophils reached >95%, as confirmed via cytospin. The purified eosinophils were resuspended in RPMI 1640 (Gibco Life Technologies, Grand Island, NY, USA) supplemented with 100 U/mL penicillin G, 10 μg/mL streptomycin, 3 μg/mL L-glutamine, and 20% heat-inactivated fetal calf serum.
Quantitative Real-Time Polymerase Chain Reaction (qRT PCR)
Eosinophils were resuspended at a concentration of 5 × 105 in 500 μL RPMI 1640 in 24-well culture plates. Eosinophils were incubated with IL-5 (25 ng/μL), IL-17A (25 ng/μL) (R&D Systems, Minneapolis, MN, USA), or phosphate-buffered saline (PBS) as a control (Sigma-Aldrich, St. Louis, MO, USA) for 30 min, 1 h, or 3 h. Cells were then harvested for total RNA isolation using the TOOLSmart RNA Extractor (BIOTOOLS CO., LTD., Taipei, Taiwan). Complementary DNA was synthesized from 1 to 100 μg of purified RNA using ToolsQuant2 Fast RT Kit (BIOTOOLS CO., LTD., Taipei, Taiwan) in accordance with the manufacturer’s instructions. We examined the mRNA expression of target genes associated with activated eosinophils, including integrin α4 for arrest on endothelium,18 integrin β2 for recruitment to airways,19 CCR3 for eotaxin receptors, and CCR5 for MIP-1 receptors20 as well as cytokine receptors, IL-5Rα, IL-17RA, IL-4Rα, prostaglandin D2 receptor 2 (CRTH2), and FcεRI, which are related to eosinophil activation or recruitment.5,6,21,22 We also examined a broader range of markers, including TGF-β1 for airway remodeling, aryl hydrocarbon receptor (AHR) for tissue adaptation under environmental triggers, and endosomal Toll-like receptors, which have been linked to eosinophil-related inflammation.23–25RT-PCR primer sets for target genes were designed manually based on the website of the National Center for Biotechnology Information using Primer-BLAST tools (see Supplementary Data, Table S1). Quantitative real-time PCR (qRT-PCR) analysis was performed using the MyGo Pro real-time PCR System (Azura Genomics Inc., Raynham, MA, USA) using 2X SuperFast SYBR qPCR Reagent (BIOTOOLS CO., LTD., Taipei, Taiwan). Ct values were derived from fluorescence measurements obtained during 3-step real-time PCR (40 cycles of denaturation at 95°C for 5 sec; annealing at 60°C for 10 sec; extension at 72°C for 15 sec). The relative gene expression was normalized to β-actin and calculated based on Ct values using PBS treatment as a baseline control. The results were expressed by 2-∆∆Ct.
Enzyme-Linked Immunosorbent Assay (ELISA)
The level of major basic protein (MBP) in the supernatant of qRT PCR 3 hr stimulation and PBS was analyzed by the ELISA kits (MyBioSource, San Diego, CA, US).
Statistical Analysis
Unpaired Student’s t-tests (two tailed) and Fisher's exact test were used to compare continuous variables and categorical data between EOA and LOA. Comparisons of mRNA expression of target genes between groups were performed using the Kruskal‐Wallis test in conjunction with the Dunn-Bonferroni post-hoc test. Comparisons of mRNA expression between different time points of stimulation were performed using Friedman test with Dunn’s mutile comparison test. mRNA expression was also tested in the context of patient atopy and obesity status (body mass index ≥30).
Spearman rank correlation was used to assess the correlations between the mRNA expression and T2 biomarkers or lung function following IL-5 or IL-17 stimulation by 30 min or 1 hour. The correlations with lung function were further analyzed using heatmaps derived using the correlation coefficient ρ. Probability values of <0.05 were considered significant. The analysis was performed using SPSS, version 19 (IBM; Armonk, NY) or Prism 8 (GraphPad software, LLC).
Results
Patient Characteristics Were Different Between LOA and EOA
This study recruited patients with EOA (n = 18) or LOA (n = 14) and healthy controls (n = 7) between October 2019 and September 2021 (Table 1). The mean age of onset was as follows: EOA (22.9 ±9.8 years) and LOA (54.7 ±12.1 years). LOA patients were older than EOA patients, were less likely to have a family history of asthma, were less prone to atopy, and presented lower FEV1/FVC levels.
 |
Table 1 Characteristics of Healthy Controls and Asthma Patients (EOA vs LOA) |
Transcriptional Activity Was Higher in Activated Eosinophils of Patients with LOA Than in Those with EOA
Figure 1 illustrates the mRNA expression of integrin α4, integrin β2, CCR3, and CCR5 (Figures 1A to D) in the eosinophils of LOA patients, EOA patients, and normal controls following stimulation using IL-5 (left panels) or IL-17 (right panels). After IL-5 stimulation for 1 hour, the increases in integrin α4 and CCR5 expression levels were more pronounced in LOA patients than in EOA patients. After IL-5 stimulation for 30 min, the increases in integrin α4, integrin β2, CCR3, and CCR5 expression levels were more pronounced in patients with LOA than in normal subjects. Following IL-5 treatment, integrin α4 expression levels were higher in EOA patients than in normal subjects. Following IL-17 stimulation for 30 min, the mRNA expression levels of all four genes were higher in LOA patients than in EOA patients and normal subjects. Figures 2 and 3 present the mRNA expression levels of the other target genes, the results of which were similar to those in Figure 1.
The results of Figure 1 – 3 are summarized in Table 2. The numerical results of mRNA expression with significant difference between groups were presented. After IL-17 stimulation, the mRNA levels of all target genes of eosinophils other than TLR-3 were higher in LOA patients than in EOA patients. We found that after IL-5 stimulation, the mRNA expression of integrin α4, CCR5, IL-5Rα, IL-17RA, CRTH2, FcεRI, TGF-β1, AHR, and TLR-9 was higher in the eosinophils of LOA patients than in those of EOA patients. Note that the effects on the mRNA expression of all genes except FcεRI and TLR-3 manifested more rapidly after IL-17 stimulation (30 min) than after IL-5 stimulation (at 1 hour). For example, the expression of integrin α4 increased higher in LOA than that of EOA after 30 min of IL-17 stimulation (2-∆∆Ct: 7.7 ± 2.1 vs 1.6 ± 0.4, p < 0.01) or 1 hour of IL-5 stimulation (2-∆∆Ct: 5.1 ± 1.0 vs 1.8 ± 1.3, p < 0.05). This implied the eosinophils of LOA were more activated than that of EOA in inflammatory milieu of asthma. IL-17 and IL-5 stimulation were both shown to increase the expression of all eosinophil target genes in LOA patients beyond that of normal controls. Note that between EOA patients and normal subjects, we observed no significant differences in the mRNA expression levels of any genes other than integrin α4.
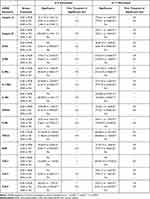 |
Table 2 Summary of Effects of IL-5 and IL-17 Stimulation on mRNA Expression in Eosinophils of EOA and LOA Patients and Normal Control Subjects |
We also checked the mRNA expression of eosinophils treated with 30 min PBS, using the formula 2−∆Ct, as the baseline mRNA expression in unstimulated eosinophils (Supplementary Data, Figure S1). By this method, we did not find a significant difference in gene expression of unstimulated eosinophils between LOA and EOA. Some genes, like CCR5, IL-17Ra, IL-4Ra, TGF-β1, TLR-3 and TLR9, expressed higher in LOA than that of normal control. However, the expression of integrin α4, CCR3, IL-5Ra, CRTH2, FcεR1, AHR and TLR-7 was higher in the normal control than LOA. These baseline values of unstimulated eosinophils cannot explain what we found in this study, the mRNA expression of target genes in eosinophils after stimulation was higher in LOA patients compared with that of EOA patients and normal control. We then compared the mRNA expression between different timepoint of stimulation (Supplementary Data, Figure S2-S15). For LOA patients, significantly higher expression of all factors was found at 30 min or 1 hour then decreased after IL-17 stimulation or IL-5 stimulation except FcεRI, AHR and TLR-3. The pattern in EOA was similar but was less pronounced. There was no difference between treatment time points in normal control subjects.
The protein level of MBP in the supernatant of rtPCR experiments after IL-5/IL-17 stimulation or BPS was no difference between EOA, LOA and NC (data hot shown).
Transcriptional Activity in Activated Eosinophils as a Function of Atopy, Obesity, or T2 Biomarkers
The mRNA expression peaked at 30 min or 1 hour after stimulation. We therefore analyzed the correlation between the mRNA levels of target genes at those timepoints versus patient characteristics.
In the EOA group after IL-5 or IL-17 stimulation for 1 hour, the mRNA expression of integrin β2 was higher in atopic patients than in non-atopic patients. In the EOA group after IL-17 stimulation for 1 hour, the mRNA expression of CRTH2 and FcεRI was higher in atopic patients than in non-atopic patients (Figure 4A).
In the EOA group after IL-17 stimulation for 30 min, the mRNA expression levels of integrin α4, CCR3, IL-5Rα, IL-17RA, CRTH2, AHR, and TLR-7 were significantly higher in obese patients than in non-obese patients (Figure 4B). In the LOA group after IL-17 stimulation for 1 hour, the mRNA expression levels of IL-17RA and TGF-β1 were higher in obese patients than in non-obese patients (Figure 4C).
Table 3 summarizes the correlation between the mRNA expression of target genes in activated eosinophils and T2 markers in patients with EOA and LOA. Among EOA patients, the mRNA expression levels of IL-17RA, CRTH2, AHR, and TLR-7 were positively correlated with total IgE levels (Spearman’s ρ was around 0.5). Among LOA patients, we observed a negative correlation between CCR5 and total IgE levels (Spearman’s ρ=−0.59). Among EOA patients, there was a negative correlation between the mRNA expression of CCR5 and TGF-β1 and eosinophil counts (Spearman’s ρ=−0.55~-0.71). Among LOA patients, we observed a positive correlation between the mRNA expression of CCR3, IL-5RA, CRTH2, AHR, and TLR-7 and eosinophil counts (Spearman’s ρ= 0.50~0.61).
 |
Table 3 mRNA Expression of Activated Eosinophil Correlation with T2 Markers in Patients with EOA and LOA |
These distinct patterns of high expression and correlation by atopy, obesity and T2 markers among EOA and LOA patients implied a complex interaction between phenotypes and eosinophil activation.
Correlation Between mRNA Gene Expression in Activated Eosinophils and Lung Function
Figure 5 presents the heatmap of the Spearman correlation coefficient ρ of mRNA expression levels and lung function parameters, where a higher ρ value indicates a stronger correlation. In the LOA group (Figure 5A), we observed a significant negative correlation between integrin α4, CCR3, CCR5, IL-5Rα, IL-17RA, CRTH2, AHR, and TLR-3, 7, and 9 mRNA expression levels and lung function in terms of FVC (L) (Spearman’s ρ=−0.56~-0.69) and FEV1 (L) (Spearman’s ρ=−0.58~-0.63). We also observed a significant negative correlation between IL-17RA, TLR-3, and TLR-9 mRNA expression levels and post-BD FEV1 (L) (Spearman’s ρ=−0.55~-0.67). TLR-3 alone was also negatively correlated with FVC (%) and FEV1/FVC. Note that all of the significant correlations between specific genes and lung function were observed only after IL-5 stimulation.
Figure 5 Continued.
As shown in Figure 5B, the correlation between mRNA expression and lung function was less pronounced among EOA patients than among LOA patients. After IL-17 stimulation, we observed a negative correlation between mRNA expression levels of some genes and changes in post-BD FEV1 changed (Spearman’s ρ=−0.48~-0.78). We also observed positive correlations between mRNA expression of some genes and lung function.
Discussion
The present study discovered that the mRNA expression of target genes associated with eosinophilic function and activation was higher and faster after stimulation by IL-5 and IL-17 in patients with LOA than in patients with EOA and normal subjects. After IL-17 stimulation, atopic and obesity status was correlated with higher transcriptional expression levels of specific genes in the eosinophils of EOA patients. We also observed the opposite correlation between T2 biomarkers and transcriptive activity of eosinophils between EOA and LOA. After IL-5 stimulation, most of the transcriptional activity in eosinophils from LOA patients was correlated with reduced lung function. To the best of our knowledge, this was the first study to reveal differences between EOA and LOA in terms of the correlation between mRNA expression patterns of activated eosinophils and clinical outcomes.
There have been relatively few studies on the patterns of eosinophilic inflammation in the context of the age of onset. One early study determined that airway eosinophilia was more common in cases of LOA than in cases of EOA and that T2 inflammatory responses were less pronounced.11 One sophisticated study based on gene set variation analysis revealed that LOA was characterized by enriched gene signatures of eosinophils.10 Note that their findings are complementary to those in the current study. Post-hoc analysis in trials of anti-IL-5 biologics for severe eosinophilic asthma revealed variations in responses as a function of the age of onset. The effects of reslizumab on exacerbation were 75% less obvious in LOA patients than in EOA patients.14 In another study on patients with frequent exacerbation, benralizumab was shown to reduce exacerbation by 70% in LOA patients and 42% in EOA patients.15 In a study conducted in Australia, the benefits of mepolizumab were more pronounced in cases involving elevated blood eosinophil levels and a later age of onset.16 These differential responses among subgroups suggest that the relative importance of eosinophilic inflammation varies as a function of age of asthma onset. In the current study, we identified a correlation between an increase in transcriptional activity in the eosinophils of LOA patients after IL-5 stimulation and poor lung function but not in that of EOA patients (Figure 5). This may provide a mechanistic explanation for the superior response of eosinophil-targeting biologics in LOA patients compared to EOA patients.
Following IL-17 treatment, we observed increases in the mRNA expression of specific genes in the eosinophils of atopic or obese EOA patients and obese LOA patients with a lesser extent (Figures 3). Note that atopy is a well-known risk factor for EOA.9 Our study revealed atopy was not to have this effect on transcriptional activity in the eosinophils of LOA patients. In one previous study, a higher BMI was significantly associated with an earlier age of onset.26 Researchers have also reported increased eosinophil uptake in the lungs of obese asthmatic patients and eosinophil accumulation in airway submucosa.27,28 Our results provide further evidence of an interaction between specific phenotypes and eosinophil activation under IL-17 stimulation in EOA patients.
The opposite of the correlation between T2 markers and transcriptive activity of eosinophils between EOA and LOA is a unique finding (Table 3). Differences in the IgE correlation patterns in EOA and LOA patients revealed that the discrepancy between allergic inflammation and eosinophil activation increased with an increase in the onset of asthma age. This result is consistent with previous findings indicating that LOA is associated with non-atopic eosinophilia.9,29 These findings also indicate that regulating the recruitment of circulating eosinophils may differ between EOA and LOA. In the case of EOA, transcriptional activity may be related to cell extravasation from circulation. In the case of LOA, it appears that transcriptional activity is related to eosinophilia or recruitment into circulation Further research will be required to obtain mechanistic explanations of these phenomena.
Our results have clinical implications pertaining to the negative correlation between the transcriptional activity of IL-5-activated eosinophils and the lower lung function of LOA patients (Figure 5A). Elevated transcriptional activity levels in eosinophils were associated with lower lung function, such as FVC and FEV1. Elevated mRNA expression levels of TLR-3 were also associated with lower FVC (%) and FEV1/FVC ratio. At present, the mechanism underlying the strong correlation between mRNA expression, eg, TLR-3 and lung function is unknown. One study on allergic rhinitis revealed that eosinophils activated by TLR3 are able to recruit leukocytes to sites of inflammation.24 Activated TLR-3 has been shown to augment the extracellular production of lung fibroblasts.30 Further mechanistic studies will be required to evaluate the activated eosinophils in LOA and lung function impairment. The correlation between mRNA and lung function is quite different between the EOA and LOA groups. The post BD FEV1 change was negatively correlated with the expression levels of specific genes after IL-17 stimulation in the EOA group (Figure 5B). This implies that the expression of these IL-17 stimulated genes is correlated with the lower bronchodilator response. This novel finding has never been reported. In a previous report, the co-expression of Th2-Th17 inflammation in asthma was characterized by high IL-17A in bronchoalveolar lavage, poor lung function, and high blood eosinophil count.31 Data obtained in the current study provide further evidence as to the importance of IL-17 in the eosinophilic inflammation in cases of EOA. Interestingly, some gene expression correlated with higher lung function, suggesting a possible protective role of eosinophils for lung function in EOA.
This study was subject to a number of limitations, which limit the generalizability of our findings. First, we observed links between mRNA expression in activated eosinophils and various features of asthma. Note, however, that identifying the mechanism by which these genes affect the characteristics of asthma was beyond the aims of the study. Second, the age and lung function of EOA and LOA patients were not balanced, which may have been confounders. Nonetheless, these patient features are similar to those reported in studies related to LOA.9,13,32–34 Besides, one previous study reported a decrease in eosinophil function among older asthma patients.35 In contrast, the increasing transcriptional level in eosinophils of LOA patients revealed by the present study is a unique finding. Third, to avoid the possibility of protein expression being skewed by in vitro cell culture conditions as mentioned in the introduction,6 we focused on the transcriptive expression. Nonetheless, the post-translational effects of gene expression are unknown and further research will be required for detecting the relevant protein expression of surface or intracellular factors.
Conclusion
This study found that patients with EOA differ considerably from those with LOA in terms of background characteristics and the mRNA expression of genes related to eosinophil activation and cell recruitment. Atopy and obesity can impact the transcriptional level of eosinophils of EOA. There is contrast correlation between T2 biomarkers and transcriptive activity of eosinophils between EOA and LOA. The high transcriptional activity in activated eosinophils and negative correlations with lung function suggest that eosinophilic inflammation plays a key role in the pathogenesis of LOA.
Abbreviations
EOA, early-onset asthma; LOA, late-onset asthma; qRT PCR, quantitative real-time polymerase chain reaction; CCR, C-C motif chemokine receptor; IL-5Rα, interleukin-5 receptor α; IL-17RA, interleukin-17 receptor A; IL-4Rα, interleukin-4 receptor α; CRTH2, prostaglandin D2 receptor 2,; FcεRI, Fcε receptor type I; TGF-β1, transforming growth factor beta 1; AHR, aryl hydrocarbon receptor; TLR, toll-like receptor.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available in the Google drive repository.
[https://drive.google.com/drive/folders/13DXdwj91b2qG4WL9LLTezFFYSNxdsj7a?usp=sharing].
Ethics Approval and Informed Consent
All patients provided written informed consent.
This study was approved by the Chang Gung Medical Foundation Institutional Review Board (No. 201900943A3).
All methods were carried out in accordance with relevant guidelines and regulations complied with the Helsinki declaration.
Funding
This study is supported by Chang-Gung Memorial Hospital (Grant no. CMRPG3J1361, CMRPG3J1362).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Volbeda F, Broekema M, Lodewijk ME, et al. Clinical control of asthma associates with measures of airway inflammation. Thorax. 2013;68(1):19–24. doi:10.1136/thoraxjnl-2012-201861
2. Green RH, Brightling CE, McKenna S, et al. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet. 2002;360(9347):1715–1721.
3. Contoli M, Baraldo S, Marku B, et al. Fixed airflow obstruction due to asthma or chronic obstructive pulmonary disease: 5-year follow-up. J Allergy Clin Immunol. 2010;125(4):830–837.
4. Bochner BS. Systemic activation of basophils and eosinophils: markers and consequences. J Allergy Clin Immunol. 2000;106(5 Suppl):S292–302.
5. Johansson MW. Activation states of blood eosinophils in asthma. Clin Exp Allergy. 2014;44(4):482–498.
6. Johansson MW. Eosinophil activation status in separate compartments and association with asthma. Front Med. 2017;4:75.
7. Koenderman L, van der Bruggen T, Schweizer RC, et al. Eosinophil priming by cytokines: from cellular signal to in vivo modulation. Eur Respir J Suppl. 1996;22:119s–125s.
8. Hakansson L, Heinrich C, Rak S, Venge P. Priming of eosinophil adhesion in patients with birch pollen allergy during pollen season: effect of immunotherapy. J Allergy Clin Immunol. 1997;99(4):551–562.
9. de Nijs SB, Venekamp LN, Bel EH. Adult-onset asthma: is it really different? Eur Respir Rev. 2013;22(127):44–52.
10. Hekking PP, Loza MJ, Pavlidis S, et al. Pathway discovery using transcriptomic profiles in adult-onset severe asthma. J Allergy Clin Immunol. 2018;141(4):1280–1290.
11. Miranda C, Busacker A, Balzar S, Trudeau J, Wenzel SE. Distinguishing severe asthma phenotypes: role of age at onset and eosinophilic inflammation. J Allergy Clin Immunol. 2004;113(1):101–108.
12. Baan EJ, de Roos EW, Engelkes M, et al. Characterization of Asthma by Age of Onset: a Multi-Database Cohort Study. J Allergy Clin Immunol Pract. 2022;10(7):1825.
13. Amelink M, de Groot JC, de Nijs SB, et al. Severe adult-onset asthma: a distinct phenotype. J Allergy Clin Immunol. 2013;132(2):336–341.
14. Brusselle G, Germinaro M, Weiss S, Zangrilli J. Reslizumab in patients with inadequately controlled late-onset asthma and elevated blood eosinophils. Pulm Pharmacol Ther. 2017;43:39–45.
15. Wenzel S, Brusselle G, Hirsch I, Zangrilli JG, Rastogi S. Benralizumab efficacy in patients with uncontrolled eosinophilic asthma by age at diagnosis. Pneumologie. 2018;52(suppl 62):PA603.
16. Harvey ES, Langton D, Katelaris C, et al. Mepolizumab effectiveness and identification of super-responders in severe asthma. Eur Respir J. 2020;55(5):546.
17. Global Initiative for Asthma. Global strategy for Asthma Management and Prevention; 2018. Available from: www.ginasthma.org.
18. Banerjee ER, Jiang Y, Henderson WR, Scott LM, Papayannopoulou T. Alpha4 and beta2 integrins have nonredundant roles for asthma development, but for optimal allergen sensitization only alpha4 is critical. Exp Hematol. 2007;35(4):605–617.
19. Johansson MW, Gunderson KA, Kelly EA, Denlinger LC, Jarjour NN, Mosher DF. Anti-IL-5 attenuates activation and surface density of beta(2)-integrins on circulating eosinophils after segmental antigen challenge. Clin Exp Allergy. 2013;43(3):292–303.
20. Oliveira SH, Lira S, Martinez AC, Wiekowski M, Sullivan L, Lukacs NW. Increased responsiveness of murine eosinophils to MIP-1beta (CCL4) and TCA-3 (CCL1) is mediated by their specific receptors, CCR5 and CCR8. J Leukoc Biol. 2002;71(6):1019–1025.
21. Cossette C, Walsh SE, Kim S, et al. Agonist and antagonist effects of 15R-prostaglandin (PG) D2 and 11-methylene-PGD2 on human eosinophils and basophils. J Pharmacol Exp Ther. 2007;320(1):173–179.
22. Rajakulasingam K, Till S, Ying S, et al. Increased expression of high affinity IgE (FcepsilonRI) receptor-alpha chain mRNA and protein-bearing eosinophils in human allergen-induced atopic asthma. Am J Respir Crit Care Med. 1998;158(1):233–240.
23. Ohno I, Nitta Y, Yamauchi K, et al. Transforming growth factor beta 1 (TGF beta 1) gene expression by eosinophils in asthmatic airway inflammation. Am J Respir Cell Mol Biol. 1996;15(3):404–409.
24. Mansson A, Fransson M, Adner M, et al. TLR3 in human eosinophils: functional effects and decreased expression during allergic rhinitis. Int Arch Allergy Immunol. 2010;151(2):118–128.
25. Mansson A, Cardell LO. Role of atopic status in Toll-like receptor (TLR)7- and TLR9-mediated activation of human eosinophils. J Leukoc Biol. 2009;85(4):719–727.
26. Holguin F, Bleecker ER, Busse WW, et al. Obesity and asthma: an association modified by age of asthma onset. J Allergy Clin Immunol. 2011;127(6):1486–1493 e1482.
27. Farahi N, Loutsios C, Tregay N, et al. In vivo imaging reveals increased eosinophil uptake in the lungs of obese asthmatic patients. J Allergy Clin Immunol. 2018;142(5):1659–1662 e1658.
28. Desai D, Newby C, Symon FA, et al. Elevated sputum interleukin-5 and submucosal eosinophilia in obese individuals with severe asthma. Am J Respir Crit Care Med. 2013;188(6):657–663.
29. Brusselle GG, Maes T, Bracke KR. Eosinophils in the spotlight: eosinophilic airway inflammation in nonallergic asthma. Nat Med. 2013;19(8):977–979.
30. Sugiura H, Ichikawa T, Koarai A, et al. Activation of Toll-like receptor 3 augments myofibroblast differentiation. Am J Respir Cell Mol Biol. 2009;40(6):654–662.
31. Irvin C, Zafar I, Good J, et al. Increased frequency of dual-positive TH2/TH17 cells in bronchoalveolar lavage fluid characterizes a population of patients with severe asthma. J Allergy Clin Immunol. 2014;134(5):1175–1186 e1177.
32. Pakkasela J, Ilmarinen P, Honkamaki J, et al. Age-specific incidence of allergic and non-allergic asthma. BMC Pulm Med. 2020;20(1):9.
33. Porsbjerg C, Lange P, Ulrik CS. Lung function impairment increases with age of diagnosis in adult onset asthma. Respir Med. 2015;109(7):821–827.
34. Westerhof GA, Vollema EM, Weersink EJ, Reinartz SM, de Nijs SB, Bel EH. Predictors for the development of progressive severity in new-onset adult asthma. J Allergy Clin Immunol. 2014;134(5):1051–1056 e1052.
35. Mathur SK, Schwantes EA, Jarjour NN, Busse WW. Age-related changes in eosinophil function in human subjects. Chest. 2008;133(2):412–419.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





