Back to Journals » Infection and Drug Resistance » Volume 15
High Levels of Methicillin-Resistant Staphylococcus aureus Carriage Among Healthcare Workers at a Teaching Hospital in Addis Ababa Ethiopia: First Evidence Using mecA Detection
Authors Desta K , Aklillu E , Gebrehiwot Y, Enquselassie F, Cantillon D, Al-Hassan L, Price JR, Newport MJ, Davey G, Woldeamanuel Y
Received 27 January 2022
Accepted for publication 28 May 2022
Published 17 June 2022 Volume 2022:15 Pages 3135—3147
DOI https://doi.org/10.2147/IDR.S360123
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Kassu Desta,1,2 Eleni Aklillu,3 Yirgu Gebrehiwot,4 Fikre Enquselassie5 ,† Daire Cantillon,6 Leena Al-Hassan,6 James R Price,6 Melanie J Newport,6 Gail Davey,5,6 Yimtubezenash Woldeamanuel1
1Department of Microbiology, Immunology, and Parasitology, School of Medicine, College of Health Sciences (CHS), Addis Ababa University (AAU), Addis Ababa, Ethiopia; 2Department of Medical Laboratory Sciences, CHS, AAU, Addis Ababa, Ethiopia; 3Division of Clinical Pharmacology, Department of Laboratory Medicine, Karolinska Institute, Karolinska, Sweden; 4Department of Obstetrics and Gynecology, School of Medicine, CHS, AAU, Addis Ababa, Ethiopia; 5School of Public Health, CHS, AAU, Addis Ababa, Ethiopia; 6Department of Global Health and Infection, Brighton and Sussex Medical School, Brighton, UK
†Fikre Enquselassie passed away on October 27, 2019
Correspondence: Kassu Desta, Tel +251 911107099, Email [email protected]
Background: Staphylococcus aureus is a major human pathogen and causes healthcare and community-acquired infection. Data on the extent of MRSA colonization among health-care workers (HCWs) in sub-Saharan Africa are limited. Hence, we determined the burden of MRSA colonisation among HCWs and administrative staff in Tikur Anbessa Specialised Hospital (TASH), College of Health Sciences (CHS), Addis Ababa University, Ethiopia.
Methods: Using a cross-sectional study design, participants were screened for MRSA colonisation between June 2018 and August 2019 using nasal swabs. The swabs were analysed using standard laboratory methods including antibiotic resistance gene, mecA. Anonymised sociodemographic data were collected by pretested questionnaires to evaluate HCWs factors associated with MRSA carriage.
Results: A total of 588 HCWs and 468 administrative staff were screened for MRSA. Women were over-represented. Overall, 49.1% (289/588) of HCWs were nurses and 25% (117/468) of the administrative staff were cleaners or laundry workers. Overall, 138 S. aureus isolates were retrieved from the nasal swabs of both groups (16.3%, 96/588 from HCWs). The burden of MRSA colonisation was 4.8% (28/580, 95% CI: 3.1– 6.5%) among HCWs compared to 0.2% (1/468, 95% CI: 0.18– 0.6%) of administrative staff (p value < 0.05). The majority of S. aureus and all MRSA isolates were resistant to penicillin. Isolates from HCWs were more resistant to tested antibiotics than administrative staff (P-value < 0.05).
Conclusion: This is the first report in Ethiopia on MRSA colonization using mecA and revealed that; (i) overall carriage rates of MRSA in HCWs are comparable with observations reported in some other countries and (ii) HCWs exhibit a higher burden of MRSA carriage than administrative staff. Our data support strategic screening of MRSA and antimicrobial stewardship for better intervention measures.
Keywords: MRSA, HCWs, administrative staff, mecA, TASH, Ethiopia
Introduction
Globally, Staphylococcus aureus is a major human pathogen that can cause a wide range of infections, both healthcare-associated and community-acquired. It also exists as a commensal and approximately 30% of the human population is colonized with S. aureus at one point in time1 and 5% are colonised with methicillin resistant S. aureus (MRSA) strains. Carriage precedes infection.2
Methicillin resistance is primarily mediated by penicillin-binding protein 2a which has a low affinity for β-lactam antibiotics. The mecA gene is part of a 21 to 60 kb Staphylococcal chromosome cassette mec (SCCmec), a mobile genetic element that may also contain genetic structures such as Tn554, pUB110, and pT181 which encode additional resistance to non-β-lactam antibiotics.3 MRSA can also acquire vancomycin resistance through a plasmid-mediated mechanism (vanA, vanB) transferred from enterococcal species.4
MRSA screening and suppression in hospitalised patients, patients undergoing surgery or haemodialysis procedures, cancer patients, neonates (especially those who are underweight) and patients in ICU, have been practiced routinely in many developed countries to minimize the prevalence and spread of MRSA infection.5,6 Health-care workers (HCWs) who are colonised with MRSA can transmit it to patients and the community. The burden of MRSA carriage among HCWs has been described in many countries around the globe.7–11 However, in low and middle-income countries including Ethiopia, there has been very little research describing carriage of MRSA among HCWs.12,13 Importantly, the few published studies use phenotypic methods to characterise MRSA, which can result in over- or underestimated prevalence rates. We aimed to determine the burden of MRSA carriage among HCWs and administrative staff at a large group of teaching hospitals in Addis Ababa, Ethiopia, using both phenotypic and genotypic methods. Moreover, we compare nasal carriage rate of HCWs and administrative staff. Knowing the real burden of MRSA will enable health-care providers and policymakers to design appropriate strategies for the control and prevention of MRSA at the study site and other similar settings.
Materials and Methods
Study Site, Design and Duration
A prospective cross-sectional study design was conducted at Tikur Anbessa Specialised Hospital (TASH), a teaching and referral hospital in Addis Ababa, Ethiopia between June and September 2018 for phenotypic analysis, while the molecular tests were performed from May to August 2019. During the data collection period, there were 1245 HCWs (400 medical doctors, 600 nurses, 70 laboratory personnel, 60 pharmacy personnel, 115 other HCWs) and 1200 administrative staff in the Hospital. All HCWs have direct patient contacts while administrative staff do not have direct roles in patient management or care. During the time of data collection CHS had two functional hospital buildings in the same compound, one administration building adjacent to the hospital and another administrative and academic campus located four kilometers away from the hospital. TASH provides medical, surgical, obstetrics and gynaecology, radiology and imaging, laboratory and pharmacy services.
Study Participants and Outcome Variables
All HCW and administrative staff working at TASH for at least 6 months prior to the data collection period were approached to join the study. Participants who had taken antimicrobial drugs in the 15 days prior to data collection, staff on leave of absence and those who have been admitted to the hospital in the month prior to data collection were excluded. The burden of MRSA from nasal swabs of study participants was the outcome variable, while sociodemographic and other work-related data were collected as independent variables.
Sample Size and Sampling Technique
The single population proportion method was used to calculate the sample size. We used a 12.7% MRSA rate from a previous study in Dessie, Ethiopia12 with a 95% confidence interval, 5% margin of error, and 10% contingency level, resulting in a sample size of 520 participants. However, we have included more HCWs than the calculated sample size to include different cadres of HCWs. The number of HCWs per cadre was allocated based on their proportion and convenience of selection. We selected administrative staff using convenience sampling until the required sample size was fulfilled.
Data Collection
Socio-demographic data were collected from HCWs using pretested self-administered questionnaires, while administrative staff were interviewed by data collectors. Sociodemographic data including age, gender-identity, information on availability of guidelines and leaflets about MRSA prevention and control, availability of adequate hand hygiene materials. Past medical and surgical history were collected both for HCWs and administrative staff.
Sample Collection and Isolate Characterisation
A nasal sample was collected from both anterior nares using a single cotton-tipped sterile moistened swab (Amie’s, Oxoid, England), placed in Amie’s transport media and transported to the laboratory in the Department of Microbiology, Immunology, and Parasitology (DMIP) at CHS, AAU for further analysis. If there was a delay, samples were kept in the refrigerator for no more than 12 hours.
All nasal swabs were cultured on mannitol salt agar or CHROMagar MRSA (Oxoid, England) and incubated overnight at 35–36°C for primary isolation of S. aureus. As per routine testing methods, colonies resembling Staphylococci species were further tested; those exhibiting catalase and coagulase, mannitol fermentation, or DNAse were identified as S. aureus. Antimicrobial susceptibility testing was determined using clinical laboratory standard institute (CLSI, 2018) methodology including, rifampin (5μg), clindamycin (2μg), trimethoprim-sulfamethoxasole (1.25/23.75μg), erythromycin (15μg), tetracycline (30μg), and penicillin (10 Units). Methicillin resistance was detected using the cefoxitin (30 μg) disc diffusion test and mecA detection. The MHA plates were incubated at 36°C for 16–18 hours, and the zone of inhibition around the disc was measured in millimetre using graduated callipers and the isolates were classified as sensitive, intermediate, or resistant according CLSI guidelines. All isolates were stored in skimmed milk glycerol medium at −30 to −70°C.14,15
Resistance Mechanism Genotyping
Genotyping was undertaken at the Department of Global Health and Infection, Brighton and Sussex Medical School, University of Sussex, UK. DNA preparation was performed using Instagene matrix solution kits (IMS, BIO-RAD, Munchen, Germany) following the manufacturer’s instructions. Briefly, 1–2 loopful of overnight growth colonies of S. aureus were suspended in a sterile Eppendorf tube and washed with sterile double distilled water (DDH2O). The pellets were suspended in 200 ul of 6% IMS and the mixture was heated in a heat block at 56°C for 20 min, then vortexed and heated for 8 min at 100°C and centrifuged at 8000x g for 2–3 minutes. The DNA quality and concentration was assessed using NanoDrop™ one Spectrophotometers (Thermo Scientific™).
PCR for mecA, vanA, and vanB Detection
The presence of mecA was amplified by polymerase chain reaction (PCR) using mecA forward and reverse primers AAAATCGATGGTAAAGGTTGGC and AGTTCTGGAGTACCGGATTTGC, respectively, following previously described methods with slight modification.14 Briefly, 22.5µl of a master mix composed of 50 units/mL of Taq DNA polymerase, 400µM of each dATP, dGTP, dCTP and dTTP, 3mM MgCl2 was mixed with 10 mmol of 0.5µl of each primer and 1.5µl of DNA product was mixed and PCR was done in a thermocycler (MRB Research, UK) with an initial 35 cycles of amplification (denaturation at 94°C for 60s, primer annealing at 60°C for 90s and primer extension at 70°C for 60s), and final extension at 72°C for 5 minutes. Then, 5μl of PCR products and 1 μL of loading dye was mixed and the band was resolved in 1.2% agarose gel prepared in 1× Tris borate EDTA (TBE) buffer containing 0.5 μg/mL of SYBER green solution. A hundred (100) bp DNA ladder was used as a molecular marker, the amplification products were electrophoresed for 1h at 100V and visualized under a gel image instrument LICOR Odyssey Fc Imager and the images saved in the computer.
vanA and vanB were amplified using PCR protocols as described elsewhere16 with slight modification. Briefly, we used the GTGACAAACCGGAGGTAATA forward and TCA CCC CTT TAA CGC TAA TA reverse primers, for vanA, and forward primer of CAG TGCATGTGCCATGGATA and reverse primer of CCG CCA TCC TCC TGC AAA AAA for vanB. A master mix (composed of 50 units/mL of Taq DNA polymerase, 400µM of each dATP, dGTP, dCTP and dTTP, 3mM MgCl2) of 22.5µl was mixed with 10 mmol of 0.5µl of each forward and reverse primers of vanA and vanB, and 1.5µl of DNA product of each gene was mixed. PCR reaction was resumed in a thermocycler (MRB Research, UK) with an initial 35 cycles of amplification (denaturation at 94°C for 60s, primer annealing at 60°C for 90s and primer extension at 70°C for 60s), and final extension at 72°C for 5 minutes. 5μl of PCR products and 1μl of loading dye were mixed and the band was resolved in 1.2% agarose gel prepared in 1× TBE buffer containing 0.5μg/mL of SYBER green solution. A hundred (100) bp DNA ladder was used as molecular marker, and the amplified products were electrophoresed for 1h at 100V. The gel was visualized under a gel image instrument, LICOR Odyssey Fc Imager and the image saved into the computer. In this study, we considered MRSA if the isolate was resistant to cefoxitin and positive for mecA.
Quality Assurance Measures
All pre-analytical, analytical, and post-analytical quality control (QC) measures were taken following standard operating procedures (SOPs) for isolation, identification, antimicrobial susceptibility testing (AST) and molecular testing. S. aureus (ATCC 25923), MRSA 252 Newman strains and E. faecium 1024 were used as QC strains. About 5% of AST was duplicated to check for any difference in performances. Known gram-positive and negative organisms were used to assess the QC of gram staining reagent, coagulase, DNAse, and catalase tests. Positive and negative controls were implemented for all PCR reactions and gel electrophoresis reading. Isolates were stored at negative 80°C with glycerol and skimmed milk.
Data Analyses
Data analyses and cleaning were done using SPSS version 20.0 software. Comparison was made for different variables among MRSA positive and negative participants and statistically tested using chi-square or Fisher’s exact test. A comparison was made between the MRSA colonisation rate of HCWs and administrative staff. A p-value of <0.05 was considered statistically significant.
Ethical Considerations
This study was conducted in accordance with the Declaration of Helsinki. Ethical approval was obtained from the Institutional Review Board of CHS, AAU (Ref. no. AAUMF 03-008) and from the national research and ethics review committee (Ref. no. MoST 310/160/18). Written informed consent was obtained from each participant. The confidentiality of all information gathered from the participants was maintained. Participants were informed on the overall findings including MRSA carriage in the hospital. The principal investigator showed photographs of MRSA cultures for HCWs, to create some awareness on MRSA and other drug-resistant bacteria.
Operational Definition and Abbreviations
Health-care workers: These are qualified health-care professionals including nurses, doctors, laboratory personnel, pharmacy personnel, radiographers and radiologists, physiotherapists, anesthetist, interns and residents who provide different types of patient care in TASH.
Methicillin resistant S. aureus: These are S. aureus isolates resistant to cefoxitin and positive for mecA.
Administrative staff: these are other than HCWs like secretary, drivers, garage workers, cleaners and laundry workers, finance and personnel officers, library staff who have no direct contacts with patients and working in TASH and CHS.
AST, Antimicrobial Susceptibility Testing; ATCC, American Type Culture Collection; CHS: College of Health Sciences, CLSI, Clinical and Laboratory Standard Institute; MRSA: methicillin resistant S. aureus; Multiple Drug Resistance; QC, Quality Control; TASH: Tikur Anbessa Specialised Hospital.
Results
Socio-Demographic Characteristics of HCWs and Administrative Staff
Overall, 1056 HCWs and administrative staff were recruited for the study; 588 were HCWs and 468 administration staff. Of the HCWs, 58.4% identified as female and the mean age was 29 years (SD ± 6.7 years, range of 20–57 years). Overall, 75% of HCWs in TASH were within the age group of 20–26 years, 40.3% (237/586) of HCWs were married; most were nurses (49.1%, 289/588) followed by medical doctors (28.4%, 167/588). In terms of educational level, 63% (369/586) of HCWs had Bachelor of Science degrees, and 60.5% (355/587) had 1–4 years of work experience in TASH (Table 1).
 |
Table 1 Socio-Demographic Characteristics and Departments of Participants in TASH, 2019 |
Of the administrative staff, 64.1% identified as female and only 4.7% (22/468) were 56 years old or above. More than 65% of participants had secondary-level education or below. Nearly 25% and 5% of them had a history of hospital admission or surgical intervention, respectively. More than 37% of the administrative staff had served for at least 8 years, and more than 57% of these were in CHS administrative offices.
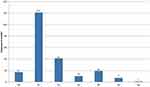
Based on our findings, 16.4% (96/585) and 15.38% (90/585) of HCWs were working in the out-patient or emergency department and Gynaecology and Obstetrics department of TASH, respectively (Figure 1).
 |
Figure 1 Distribution of HCWs in the various departments of TASH, 2019, the bar indicate the proportion of HCWs in each department as showed in the y-axis and x –axis is the proportion (%). |
Importantly, 86.1% (504/586) and 84.6% (496/190/586) of HCWs at TASH said that there was no MRSA-related guideline or leaflets, and insufficient hand hygiene materials in TASH, respectively, while 88% (515/585) had not had training on MRSA prevention and control.
Among the HCWs in TASH, 82.5% (484/587) had no history of hospital admission in the 3 years prior to data collection, and only 11.5% (67/585) had surgical intervention in the hospital in the same period. Of the 468 administrative staff, 25% (117/468) were cleaners and laundry workers (Figure 2). Only 24.6% (115/468) and 5.1% (24/468) had a hospital admission or surgical intervention in the 3 years prior to data collection.
The Isolation Rate of S. aureus, and Antimicrobial Susceptibility Testing Patterns
The proportion of S. aureus isolates from nasal swabs of HCWs and administrative staff was 16.3% (96/588) and 9.1% (42/468), respectively. All 138 S. aureus isolates underwent AST (Table 2). Among S. aureus isolates, 21.7% were susceptible to penicillin and all isolates were susceptible to rifampicin. Twenty-nine isolates were MRSA as characterised by resistance to cefoxitin (oxacillin) and contained mecA.
 |
Table 2 Antimicrobial Susceptibility Patterns of S. aureus Isolates from Nasal Swabs of HCWs and Administrative Staff of TASH, CHS, AAU, Ethiopia |
In this study, both MSSA and all MRSA isolates were tested for vancomycin and all of them were sensitive using the molecular method.
MRSA strains had higher resistance rate to most drugs tested than MSSA isolates (p-value <0.05).
Multidrug-Resistant Pattern of S. aureus Isolates
One hundred and twenty-one (87.6%) of S. aureus isolates were resistant to at least one antibiotic, 41 isolates were resistant to two different classes of antibiotics, and one isolate was resistant to six antibiotics (Figure 3).
MRSA Burden Among HCWs and Administrative Staff
The burden of MRSA nasal colonisation among HCWs was 4.8% (95% CI: 3.1–6.5%, 28/580) and was comparable among those identifying as male and female (4.9% and 4.7%, respectively, p-value 0.517). Figure 4 shows the amplified mecA.
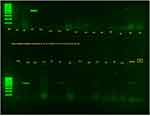 |
Figure 4 Gel band result of mecA. MM is for molecular markers of 100 bp; the letters from D7 to F10 are PCR products of S. aureus isolates; MRSA is a positive control and NC is a negative control. |
The burden of MRSA was higher in HCWs within the age group of 20–26 years (Table 3). Nurses had the highest MRSA rate in TASH (22/28 from the total positive).
 |
Table 3 Nasal Colonisation of HCWs with MRSA Working at TASH, CHS, AAU, 2019 |
Most of the HCWs had 1–2 years of work experience and nasal MRSA colonisation was found in 6.59% (13/197). MRSA colonisation of HCWs did not differ according to - whether HCWs had had MRSA-related training or not; availability of guidelines; source of information; history of hospital admission; surgical intervention; availability of sufficient hand hygiene materials; or department in which they were working (P-value >0.05; data not shown).
On the other hand, among 468 administrative staff, only one individual had MRSA carriage in the nasal cavity, with a magnitude of 0.2% (95% CI: 0.18–0.6%, 1/468). The MRSA-positive administrative staff member was a 26-year-old female cleaner with work experience of 4 years, but no history of hospital admission or surgical intervention for the last three years during data collection time.
Discussion
MRSA-colonised patients and HCWs could increase the risk of transmission and contamination of the hospital environment. Screening of patients and HCWs has been implemented in some hospital settings.17 HCWs are on the boundary between the community and hospital, and hence they may be at increased risk of MRSA colonisation or infection.18
In the current study, higher rate of S. aureus carriage was seen among HCWs than administrative staff (16.3% and 9.0% respectively) which is slightly lower than two previous studies done in Mekele and Adigrat/Wukro hospitals of Ethiopia (28.8% and 20.3% respectively).13,19 Higher rate of S. aureus carriage was also reported from Uganda (48%) and Gaza (25.5%).20,21 The discrepancies seen could be explained as there is difference in sample size, laboratory methods and difference in study sites.
The 78.2% penicillin resistance in this study was comparable with the 71% resistance reported in the Netherlands22 but lower than two former studies done in Dessie and Adigrat, Ethiopia (100% resistant)19,23 and 90.6% to 100% in Southern Brazil and Gaza.21,24 The rate of multidrug-resistance was similar to reports from Dessie and Jimma, Ethiopia.23,25
The rate of resistance to cefoxitin (oxacillin) in this work (21%) was higher than two previous studies in Ethiopia namely Adigrat/Wukro hospital (5.8%) and Dessie Hospitals (12.7%).19,23 This might be due to the higher number and diversity of the sources of our isolates and the use of oxacillin as a surrogate marker in the aforementioned studies.
Erythromycin resistance in this study (18.1%) is lower than 29–100% resistance rates reported from India, Nigeria and Libya.26–28 These differences in availability and antibiotics use in the study sites could contribute the inconsistency in the resistance rate.
Resistance rate of 12.3% (17 isolates) to clindamycin in this study was lower than an earlier study in North Ethiopia (17.2%).19 Clindamycin resistance was higher for strains isolated from HCWs than administrative staff isolates, indicating that exposure to resistant strains is more likely in the hospital setting than in the community. Higher resistance was also reported from studies from Libya and India (15–69%).26,28
Resistance to tetracycline (36.2%) and trimethoprim-sulfamethoxazole (68.9%) in our study was comparable with the findings from North Ethiopia (55.2% and 51.7% respectively).19 However, a higher resistance rate to tetracycline (86.1%) was reported from Mekele, North Ethiopia.13 Probably our isolates mainly from administrative staff could comprise susceptible strains and contribute to low levels of resistance to this drug.
One of the encouraging results in our study is that, all isolates were sensitive to rifampicin and vancomycin, which is in line with studies in Nepal29 and Southern Brazil,24 suggesting these two drugs are still useful and effective for managing S. aureus and MRSA-related infections. Although the source population was different, a recent study in North Ethiopia also indicated the absence of resistance to these drugs.30 Resistance to rifampin was reported among 50% of MRSA and 6% of MSSA in one of the Indian studies.26 Since rifampicin is not routinely used for the treatment of S. aureus infection in Ethiopia, this might favour the absence or selection of resistant strains.
However, 5.6% rate of resistance to vancomycin was reported in a former study in Mekele, Ethiopia,13 rate of 1.3% and 6% in Nigerian studies,27,31 and 12% in Libya28 were also seen. We have used PCR for the detection of vanA and vanB unlike the other studies in Ethiopia, Nigeria and Libya that used only the disc diffusion test and may contribute the existence of vancomycin resistance.
Generally, we found higher rate of drug resistance for MRSA than MSSA isolates and among HCWs than administrative staff (p-value <0.05), which is in line with other studies.26,32,33 This finding underscores that HCWs are exposed to more resistant strains than administrative staff who had no or very limited contact with patients.
The 4.8% (28/580) MRSA rate in our HCWs is in line with the figure of 4.6% from a meta-analysis of 127 studies around the world,18 rate of 1.8–4.4% from a systematic review in Europe and the USA,34 and of 4.6% from a meta-analysis in nine German acute care hospitals35 in a non-outbreak setting. However, our MRSA burden is lower than previous studies done in Ethiopia with a rate of 5.8% to 14.1%,12,13,20,25,36 and elsewhere with MRSA rates of 11.7% to 52.59%.21–28,37 The higher rate might be, other studies used resistant testing based on disc diffusion alone and so overestimate the MRSA rate, whereas we used cefoxitin disc diffusion and mecA methods together. Moreover, our sample size was reasonably large and includes diverse cadres of HCWs.
Lower MRSA burdens than ours have been reported elsewhere, with MRSA rates of 0% to 2.3% in Kenya, Germany, Australia, and Nigeria.27,38–40 Better infection prevention practices in Europe and the involvement of HCWs who had very low or limited contact with patients in Kenyan and Nigerian studies might have favored for the lower MRSA colonization rates.
Almost all Ethiopian studies reported higher rates of MRSA carriage than ours, as they used coagulase tests alone for identification purposes, rather than coagulase and DNAse tests. These findings need further investigation as we have seen discrepancies in S. aureus identification that could potentially result in inflated rates of MRSA.41 This is expected in low-income settings where the microbiology laboratory infrastructure and personnel are not well supported. We feel that there might be over-reporting of MRSA in most of the previous studies done in Ethiopia and other similar settings around the globe.
Earlier work from Ethiopia and elsewhere9,12,13,21,22,26,27,33,37 indicated that MRSA rates might be higher among nursing staff, in agreement with this study. This may be the consequence of more frequent contact with patients and hence exposed them for MRSA.
In the current study, the majority of MRSA carriage (92.8%; 26/28) was seen among nurses and HCWs in the age group of 20–26 years, which is similar to other studies.29,31 This age group is active and may have more interaction with patients and colleagues, potentially exposing them to more MRSA colonization. As HCWs become more senior, they may have less contact with patients. Other variables like educational, marital status, work experience, history of hospital admission and surgical procedure were not statistically associated with MRSA burden in line with previous studies in Ethiopia.12,13
All in all, this study generates robust evidence on the burden of MRSA using phenotypic and genotypic methods which are very rarely done in Ethiopia and other sub-Saharan Africa. Our work also corroborates that, HCWs had higher rates of MRSA. As a limitation, we did not analyse a second swab after an interval of time to understand the persistence of MRSA carriage. Second, SCCmec typing was not done which could predict the source of MRSA. Finally, we did not perform co-colonisation of MRSA and MSSA among our study participants which might be addressed in future works.
Conclusion
MRSA carriage was much higher among HCWs of TASH than administrative staff. Importantly, vancomycin and rifampicin are still the most effective antibiotics. Updating the real burden of MRSA carriage using phenotypic and genotypic methods is essential to inform preventive measures and track changes for intervention in TASH and other similar settings. Moreover, TASH being as a tertiary level specialised teaching hospital should have emerging technologies including molecular identification methods, drug resistance testing and strategic screening of HCWs for MRSA. Although we have generated evidence on the burden of MRSA carriage at TASH, it is difficult to recommend decolonisation of HCWs without acquiring additional evidences.
Data Sharing Statement
We have presented the most important data with this report. However, upon reasonable request, additional data could be available from the corresponding author.
Acknowledgments
The authors wish to thank Addis Ababa University, the Department of Global Health and Infection at Brighton and Sussex Medical School, and CDT Africa for financial support. We thank also all study participants and colleagues for their willingness and support of this study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article was submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors declared no conflicts of interest in relation to this work.
References
1. Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661. doi:10.1128/CMR.00134-14
2. Calfee DP, Salgado CD, Milstone AM, et al. Strategies to prevent methicillin-resistant Staphylococcus aureus transmission and infection in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol. 2014;35(7):772–796. doi:10.1086/676534
3. Zaghloul MZ. Methicillin-resistant Staphylococcus aureus (MRSA). J Med Microb Diagn. 2016;5:2. doi:10.4172/2161-0703.1000e131
4. Shariati A, Dadashi M, Moghadam MT, van Belkum A, Yaslianifard S, Darban-Sarokhalil D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10(12689). doi:10.1038/s41598-020-69058-z
5. Chen AF, Wessel CB, Rao N. Staphylococcus aureus screening and decolonization in orthopaedic surgery and reduction of surgical site infections. Clin Orthop Relat Res. 2013;471:2383–2399. doi:10.1007/s11999-013-2875-0
6. Rao N, Cannella B, Crossett LS, Yates AJ Jr, McGough R, A preoperative decolonization protocol for Staphylococcus aureus prevents orthopaedic infections. Clin Orthop Relat Res. 2008;466:1343–1348. doi:10.1007/s11999-008-0225-4
7. Drago L, Cappelletti L, Lamartina C, Berjano P, Mattina R, De Vecchi E. Colonization by methicillin resistant Staphylococci of nares and skin in healthcare workers: a pilot study in spinal surgeries. Injury. 2015;46(S8):S77–S80. doi:10.1016/S0020-1383(15)30059-0
8. Maheshwari M, Devi S, Agarwal P, Malhotra VL. Screening of health care workers for nasal and hand carriage of multi-drug resistant organisms in a teaching hospital in Rural Haryana, India. IJSR. 2014;3(11):369–371.
9. Knahal R, Sah P, Lamichhane P, Lamsal A, Upadhaya S, Pahwa VK. Nasal carriage of methicillin resistant Staphylococcus aureus among health care workers at a tertiary care hospital in Western Nepal. Antimicrob Resist Infect Control. 2015;4:39. doi:10.1186/s13756-015-0082-3
10. Verwer PE, Robinson JO, Coombs GW, et al. Prevalence of nasal methicillin-resistant Staphylococcus aureus colonization in healthcare workers in a Western Australian acute care hospital. Eur J Clin Microbiol Infect Dis. 2012;31:1067–1072. doi:10.1007/s10096-011-1408-6
11. Amorim ML, Vasconcelos C, Oliveira DC, et al. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization among patients and healthcare workers in a Portuguese hospital: a pre-intervention study toward the control of MRSA. Microb Drug Resist. 2009;15(1):19–26. doi:10.1089/mdr.2009.0881
12. Shibabaw A, Abebe T, Mihret A. Nasal carriage of methicillin resistant Staphylococcus aureus among Dessie Referral Hospital health care workers; Dessie, Northeast Ethiopia. Antimicrob Resist Infect Control. 2013;2:25. doi:10.1186/2047-2994-2-25
13. Gebreyesus A, Gebre-Selassie S, Mihert A. Nasal and hand carriage rate of methicillin resistant Staphylococcus aureus (MRSA) among health care workers in Mekelle Hospital, North Ethiopia. Ethiop Med J. 2013;51(1):41–47.
14. Price JR, Cole K, Bexley A, et al.; The Modernising Medical Microbiology Informatics Group. Transmission of Staphylococcus aureus between health-care workers, the environment, and patients in an intensive care unit: a longitudinal cohort study based on whole-genome sequencing. Lancet Infect Dis. 2017;17:207–214.
15. Clinical and Laboratory Standards Institute (CLSI, 2018). Methods for Dilution of Antimicrobial Susceptibility Testing for Bacteria That Grow Aerobically.
16. Saadat S, Solhjoo K, Norooz-Nejad MJ, Kazemi A. VanA and VanB positive vancomycin-resistant Staphylococcus aureus among clinical isolates in Shiraz, South of Iran. Oman Med J. 2014;29(5):335–339. doi:10.5001/omj.2014.90
17. Albrich WC, Harbarth S. Healthcare workers: source, vector or victim of MRSA. Lancet Infect Dis. 2008;8:289–301. doi:10.1016/S1473-3099(08)70097-5
18. Hadley S, Immerman I, Hutzler L, Slover J, Bosco J. Staphylococcus aureus decolonization protocol decreases surgical site infections for total joint replacement. Arthritis. 2010;2010:1–4. doi:10.1155/2010/924518
19. Legesse H, Kahsay AG, Kahsay A, et al. Nasal carriage of, risk factors and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus among health care workers in Adigrat and Wukro hospitals, Tigray, Northern Ethiopia. BMC Res Notes. 2018;11(1):250. doi:10.1186/s13104-018-3353-2
20. Abimana JB, Kato CD, Bazira J. Methicillin resistant Staphylococcus aureus nasal colonization among health care workers at Kampala International University Teaching Hospital, Southwestern Uganda. Can J Infect Dis Med Microbiol. 2019;2019. doi:10.1155/2019/4157869.
21. El Aila NA, Al Laham NA, Ayesh BM. Nasal carriage of methicillin resistant Staphylococcus aureus among health care workers at Al Shifa hospital in Gaza Strip. BMC Infect Dis. 2017;17:28. doi:10.1186/s12879-016-2139-1
22. Rijnders MIA, Nys S, Driessen C, et al. Staphylococcus aureus carriage among GPs in the Netherlands. Br J Gen Pract. 2010;60:902–906. doi:10.3399/bjgp10X544078
23. Shibabaw A, Abebe T, Mihret A. Antimicrobial susceptibility pattern of nasal Staphylococcus aureus among Dessie Referral Hospital health care workers, Dessie, Northeast Ethiopia. Int J Infect Dis. 2014;25:22–25. doi:10.1016/j.ijid.2014.03.1386
24. Danelli T, Duarte FC, de Oliveira TA, et al. Nasal carriage by Staphylococcus aureus among healthcare workers and students attending a University Hospital in Southern Brazil: prevalence, phenotypic, and molecular characteristics. Interdiscip Perspect Infect Dis. 2020;2020:11. doi:10.1155/2020/3808036
25. Efa F, Alemu Y, Beyene G, Gudina EK, Kebede W. Methicillin resistant Staphylococcus aureus carriage among medical students of Jimma University, Southwest Ethiopia. Heliyon. 2019;5(1):e01191. doi:10.1016/j.heliyon.2019.e01191
26. Agarwal L, Singh AK, Sengupta C, Agarwal A. Nasal carriage of methicillin- and mupirocin-resistant S. aureus among health care workers in a tertiary care hospital. J Res Pharm Pract. 2015;4:182–186. doi:10.4103/2279-042X.167046
27. Fadeyi A, Bolaji B, Oyedepo O, et al. Methicilin resistant Staphylococcus aureus carriage amongst healthcare workers of the critical care units in a Nigerian Hospital. Am J Infect Dis. 2010;6(1):18–23. doi:10.3844/ajidsp.2010.18.23
28. Ahmed MO, Elramalli AK, Amri SG, Abuzweda AR, Abouzeed YM. Isolation and screening of methicillin-resistant Staphylococcus aureus from health care workers in Libyan hospitals. EMHJ. 2012;18(1):37–42. doi:10.26719/2012.18.1.37
29. Khatri S, Pant ND, Bhandari R, et al. Nasal carriage rate of methicillin resistant Staphylococcus aureus among health care workers at a Tertiary Care Hospital in Kathmandu, Nepal. J Nepal Health Res Counc. 2017;15(1):26–30. doi:10.3126/jnhrc.v15i1.18009
30. Kalayu AA, Woldetsadik DA, Woldeamanuel Y, Wang SH, Gebreyes WA, Teferi T. Burden and antimicrobial resistance of S. aureus in dairy farms in Mekelle, Northern Ethiopia Alem. BMC Vet Res. 2020;16:20. doi:10.1186/s12917-020-2235-8
31. Nwokah EG, Eddeh-Adjugah O, Aleru CP. Assessment of asymptomatic methicillin resistant Staphylococcus aureus carriage among health care workers in the University of Port Harcourt teaching Hospital, Nigeria. SCIREA J Health. 2017;2(2). Available from: http://www.scirea.org/journal/PMH.
32. Walana W, Bobzah BP, Kuugbee ED, et al. Staphylococcus aureus nasal carriage among healthcare workers, inpatients and caretakers in the Tamale Teaching Hospital, Ghana. Sci Afr. 2020;8:e00325. doi:10.1016/j.sciaf.2020.e00325
33. Okamo B, Moremi N, Seni J, Mirambo MM, Kidenya BR, Mshana SE. Prevalence and antimicrobial susceptibility profiles of Staphylococcus aureus nasal carriage among pre-clinical and clinical medical students in a Tanzanian University. BMC Res. 2016;9:47. doi:10.1186/s13104-016-1858-0
34. Dulon M, Peters C, Schablon A, Nienhaus A. MRSA carriage among healthcare workers in non-outbreak settings in Europe and the United States: a systematic review. BMC Infect Dis. 2014;14:363. doi:10.1186/1471-2334-14-363
35. Sassmannshausen R, Deurenberg RH, Köck R, et al. MRSA prevalence and associated risk factors among health-care workers in non-outbreak situations in the Dutch-German EUREGIO. Front Microbiol. 2016;7:1273. doi:10.3389/fmicb.2016.01273
36. Reta A, Mengist A, Tesfahun A. Nasal colonization of methicillin resistant Staphylococcus aureus in Ethiopia: a systematic review and meta-analysis. Ann Clin Microbiol Antimicrob. 2019;18:25. doi:10.1186/s12941-019-0324-y
37. Joachim A, Moyo SJ, Nkinda L, et al. Nasal carriage of methicillin-resistant Staphylococcus aureus among health care workers in tertiary and regional hospitals in Dar es Salam, Tanzania. Int J Microbiol. 2018;2018:7. doi:10.1155/2018/5058390
38. Omuse G, Kariuki S, Revathi G. Unexpected absence of methicillin resistant Staphylococcus aureus nasal carriage by health care workers in a tertiary hospital in Kenya. J Hosp Infect. 2012;80(1):71–73. doi:10.1016/j.jhin.2011.09.009
39. Schubert M, Kämpf D, Wahl M, et al. MRSA point prevalence among health care workers in German rehabilitation centers: a multi-center, cross-sectional study in a non-outbreak setting. Int J Environ Res Public Health. 2019;16(9):1660. doi:10.3390/ijerph16091660
40. Munckhof WJ, Nimmo GR, Schooneveldt JM, et al. Nasal carriage of Staphylococcus aureus, including community-associated methicillin-resistant strains, in Queensland adults. Clin Microbiol Infect. 2009;15(2):149–155. doi:10.1111/j.1469-0691.2008.02652
41. Becker K, Pagnier I, Schuhen B, et al. Does Nasal Co-colonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J Clin Microbiol. 2006;44(1):229–231. doi:10.1128/JCM.44.1.229-231.2006
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.