Back to Journals » Cancer Management and Research » Volume 13
Grape Seed Proanthocyanidins (GSPs) Inhibit the Development of Cutaneous Squamous Cell Carcinoma by Regulating the hsa_circ_0070934/miR-136-5p/PRAF2 Axis
Authors Xiong W, Wu L, Tang R, Zhang Q, Guo Q, Song S
Received 14 January 2021
Accepted for publication 26 April 2021
Published 1 June 2021 Volume 2021:13 Pages 4359—4371
DOI https://doi.org/10.2147/CMAR.S302084
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Weibiao Xiong,1,* Lan’e Wu,1,* Runke Tang,2 Qingqing Zhang,1 Qian Guo,1 Shuhua Song3
1Xiong Wei-Biao Workroom, Jiangxi Province Hospital of Integrated Chinese and Western Medicine, Nanchang, Jiangxi, 330003, People’s Republic of China; 2Department of Rehabilitation, Jiangxi Province Hospital of Integrated Chinese and Western Medicine, Nanchang, Jiangxi, 330003, People’s Republic of China; 3Department of Dermatology, Jiangxi Province Hospital of Integrated Chinese and Western Medicine, Nanchang, Jiangxi, 330003, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lan’e Wu
Xiong Wei-Biao Workroom, Jiangxi Province Hospital of Integrated Chinese and Western Medicine, No. 90, Bayi Road, Nanchang, Jiangxi, 330003, People’s Republic of China
Tel +86-0791-86292774
Email [email protected]
Background: Grape seed proanthocyanidins (GSPs) have been shown to inhibit the progression of many cancers, including cutaneous squamous cell carcinoma (CSCC). Circular RNA (circRNA) is a key regulator for cancer progression. However, it is unclear whether GSPs can mediate the progression of CSCC by regulating circRNA.
Methods: Quantitative real-time PCR was conducted to determine the expression of hsa_circ_0070934, microRNA (miR)-136-5p and prenylated Rab acceptor family 2 (PRAF2). MTT assay and colony formation assay were used to assess cell proliferation. Cell cycle process and apoptosis were detected by flow cytometry, and cell migration and invasion were measured by transwell assay. Western blot analysis was utilized to examine protein expression. In addition, dual-luciferase reporter assay and RIP assay were used to evaluate the interaction between miR-136-5p and hsa_circ_0070934 or PRAF2. Subcutaneous xenograft models were constructed to explore the function of GSPs on CSCC tumor growth in vivo.
Results: GSPs could reduce hsa_circ_0070934 expression and inhibit CSCC cell proliferation, cell cycle process, migration, invasion, while promote apoptosis. Overexpressed hsa_circ_0070934 could reverse the suppressive effect of GSPs on CSCC cell progression. MiR-136-5p could be sponged by hsa_circ_0070934, and its overexpression also abolished the positively regulation of hsa_circ_0070934 on the progression of GSPs-induced CSCC cells. PRAF2 was a target of miR-136-5p, and its expression could be decreased by GSPs and increased by hsa_circ_0070934. The inhibitory effect of miR-136-5p on CSCC cell progression could be reversed by PRAF2 overexpression. Additionally, GSPs also could inhibit CSCC tumor growth in vivo.
Conclusion: Our data showed that GSPs regulated the hsa_circ_0070934/miR-136-5p/PRAF2 axis to restrain CSCC progression.
Keywords: cutaneous squamous cell carcinoma, GSPs, hsa_circ_0070934, miR-136-5p, PRAF2
Introduction
Cutaneous squamous cell carcinoma (CSCC) is a kind of malignant tumor caused by excessive proliferation of skin epidermis or keratinocytes, which is a common skin cancer.1,2 Ultraviolet radiation, radiation damage, and chemical irritation are considered to be important causes of CSCC.3,4 Due to the high degree of malignancy, the treatment options for locally advanced CSCC or metastatic CSCC are very limited, which often leads to poor prognosis for patients.5,6 Therefore, finding an effective way to treat CSCC is essential to provide further guidance in the clinic.
Grape seed proanthocyanidins (GSPs) are an effective natural antioxidant that can scavenge free radicals in the human body. A large number of studies have shown that GSPs have a wide range of nutritional value and health care effects,7,8 and also have a better inhibitory effect on the development of many cancers, including pancreatic cancer and cervical cancer.9,10 In CSCC, Sun et al suggested that GSPs could suppress the invasion of head and neck CSCC.11 Hence, GSPs might be an effective natural extract to inhibit the progression of CSCC.
As a special non-coding RNA, the important role of circular RNA (circRNA) in cancer has been clarified. CircRNA is proved to have a key regulatory function on the malignant progression of cancer, and can be used as a biomarker for cancer treatment and diagnosis.12,13 For example, hsa_circ_101996 could increase the proliferation and invasion of cervical cancer,14 while hsa_circ_103809 had an inhibition on colorectal cancer proliferation and migration.15 Hsa_circ_0070934 was a newly discovered circRNA with significantly high expression in CSCC, which had been confirmed to be associated with the proliferation, metastasis and apoptosis of CSCC.16,17 Therefore, hsa_circ_0070934 might be a key target to regulate CSCC progression.
In our research, we found that GSPs could decrease the expression of hsa_circ_0070934 in CSCC cells, so we proposed the hypothesis that GSPs mediated the progression of CSCC by regulating hsa_circ_0070934. In addition, many evidences indicate that circRNA can act as a sponge of microRNA (miRNA) to participate in the regulation of downstream genes.18,19 Further research proposed the existence of the circRNA/miRNA/mRNA axis, which perfected the potential molecular mechanism of GSPs regulating CSCC progression.
Materials and Methods
Cell Culture and GSPs Treatment
Human CSCC cell lines (A431 and SCL-1) and normal epidermal cells (HaCaT) were obtained from Biovector National Typical Culture Center (NTCC, Beijing, China). All cells were cultured in DMEM medium (Gibco, Grand Island, NY, USA) at 37°C with 5% CO2 incubator. Fetal bovine serum (FBS, 10%, Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA) were added to the DMEM medium to prepare the complete medium. GSPs (Chengdu Must Bio-technology Co., Ltd., Sichuan, China) were dissolved in DMSO (Solarbio, Beijing, China) and prepared to different concentrations (10, 20 and 40 μg/mL). When the cells reached 80% confluences, the cells were treated with different concentrations of GSPs for 24 h. Cells were treated with an equal amount of DMSO as negative control, denoted as 0 μg/mL GSPs.
Cell Transfection
A431 and SCL-1 cells were seeded into 6-well plates and cultured until the cell density reached 50–60%. Cell transfection was performed using Lipofectamine 3000 (Invitrogen). All vectors and oligonucleotides were synthesized from RiboBio (Guangzhou, China), including hsa_circ_0070934 overexpression vector and small interference RNA (hsa_circ_0070934 and si-hsa_circ_0070934) or their negative controls (Vector and si-NC), miR-136-5p mimic and inhibitor or their negative controls (NC mimic and NC inhibitor), pcDNA prenylated Rab acceptor family 2 (PRAF2) overexpression vector and its control (pcDNA). After transfection for 24 h, the cells were treated with 20 μg/mL GSPs for 24 h. After that, the cells were collected for functional experiments.
Quantitative Real-Time PCR (qRT-PCR)
TRIzol reagent (Invitrogen) was used to extract RNA and SuperScrip VILO™ cDNA Synthesis Kit (Invitrogen) was used to reverse-transcript RNA to cDNA. Then, qRT-PCR was conducted using SYBR Green (Solarbio) in PCR system. Relative expression was normalized to β-actin or U6 and calculated with 2−ΔΔCt method. The sequences of primers were listed as follows: hsa_circ_0070934, F 5ʹ-GGGTGGTAATATCCGAGGTTCC-3ʹ, R 5ʹ-TTGTCTTGAGCTTTCCTGCCT-3ʹ; miR-136-5p, F 5ʹ-GCCGAGACTCCATTTGTTTTGAT-3ʹ, R 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ; PRAF2, F 5ʹ-CTGGACGACTTTGTTCTGGGG-3ʹ, R 5ʹ-GCTCAGGAGCGTATGAAGTGG-3ʹ; GAPDH, F 5ʹ-AAGGCTGTGGGCAAGGTCATC-3ʹ, R 5ʹ-GCGTCAAAGGTGGAGGAGTGG-3ʹ; β-actin, F 5ʹ-ATAGCACAGCCTGGATAGCAACGTAC-3ʹ, R 5ʹ-CACCTTCTACAATGAGCTGCGTGTG-3ʹ; U6, F 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ, R 5ʹ-GGAACGCTTCACGAATTTG-3ʹ.
Identification of circRNA
The PCR product of hsa_circ_0070934 was sequenced. After genomic DNA (gDNA) was obtained using Universal gDNA Extraction Kit (Takara), the cDNA and gDNA of hsa_circ_0070934 and GAPDH were amplified with convergent primers and divergent primers. The collected PCR products were used for agarose gel electrophoresis.
MTT Assay
After transfected or treated, A431 and SCL-1 cells were harvested and then re-plated into 96-well plates (2 × 103 cells/well) and cultured until the cells were attached to the bottom of the plate. After incubating for 0, 1, 2 and 3 days, MTT solution (Beyotime, Shanghai, China) was added to cells for 4 h and then Formazan solution was added to cells for 2 h. The optical density (OD) value was measured at 570 nm using a microplate reader (BioTek, Burlington, VT, USA).
Colony Formation Assay
After transfected or treated, A431 and SCL-1 cells were harvested and then re-seeded into 6-well plates (150 cells/well). After culturing for 2 weeks, the colonies were fixed with methanol (Chron Chemicals, Qionglai, China) and stained with crystal violet solution (Beyotime). Under a microscope (Olympus, Tokyo, Japan), the colonies were photographed and their number was counted.
Flow Cytometry
Cell cycle process and cell apoptosis were measured by this assay. After transfected or treated, A431 and SCL-1 cells were treated with trypsin (Gibco) to collect the cell suspensions (1 × 106 cells). According to the protocols of Cell Cycle Analysis Kit (Beyotime), the cell suspensions were fixed with 70% ethanol at 4°C overnight. After that, the cells were stained with Propidium Iodide and RNase A for 30 min. Cell cycle distribution was analyzed by a flow cytometer (BD Bioscience, San Jose, CA, USA). In addition, according to the protocols of Annexin V-FITC Apoptosis Detection Kit (Beyotime), the cell suspensions were suspended with binding buffer and then stained with Annexin V-FITC and Propidium Iodide. Under a flow cytometer, the apoptotic cells were analyzed.
Transwell Assay
After transfected or treated, A431 and SCL-1 cells were collected and resuspended with serum-free medium, and then seeded in the upper chamber of 24-well Transwell chamber (BD Bioscience) (2 × 105 cells/well for cell migration and 4 × 105 cells/well for cell invasion). The chambers pre-coated with a Matrigel (BD Bioscience) were used to detect cell invasion, and non-coated chambers were used to measure cell migration. Next, complete medium was added to the lower chamber. After 24 h, the cells that migrated and invaded to the bottom of the chambers were fixed with methanol and stained with crystal violet solution, and then their number was counted under a microscope (Olympus, 100×).
Western Blot (WB) Analysis
Total protein was isolated using RIPA lysis buffer (Beyotime). Then, the protein sample was electrophoresed on 10% SDS-PAGE gel and electro-transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). After blockage with skimmed milk, the membrane was incubated with primary and secondary antibodies one by one. The primary antibodies were as follows: anti-proliferating cell nuclear antigen (anti-PCNA, 1:2000, Boster, Wuhan, China), anti-caspase 3 (1:1000, Beyotime), anti-MMP-3 (1:2000, Boster), anti-PRAF2 (1:3000, Boster) and anti-β-actin (1:10,000, Boster). The secondary antibody was HRP-conjugated AffiniPure goat anti-rabbit IgG (H+L) (1:10,000, Boster). The signal was developed using BeyoECL Moon ECL Chemiluminescence Kit (Beyotime).
Dual-Luciferase Reporter Assay
According to the binding sites of miR-136-5p in hsa_circ_0070934 or PRAF2 3ʹUTR, the wild-type (WT) and mutant-type (MUT) reporter vectors of hsa_circ_0070934 or PRAF2 3ʹUTR were constructed using pGL3 reporter vector (Promega, Madison, WI, USA). The reporter vectors were co-transfected into A431 and SCL-1 cells with miR-136-5p mimic or NC mimic. After transfection for 48 h, Dual Luciferase Reporter Gene Assay Kit (Beyotime) was used for measuring relative luciferase activity.
RIP Assay
EZ-Magna RIP Kit (Millipore) was utilized for RIP assay. Briefly, A431 and SCL-1 cells were lysed with RIP buffer, and then the cell lysates were incubated with Protein A/G magnetic beads conjugated with anti-AGO2 (Millipore) or anti-IgG (Millipore). After washing with Proteinase K, the immunoprecipitated RNAs were isolated for detecting the enrichment of hsa_circ_0070934, miR-136-5p and PRAF2 using qRT-PCR.
Subcutaneous Xenograft Models
Ten male BALB/c nude mice (5 weeks old) were obtained from Vital River (Beijing, China). All animal protocols were approved by Jiangxi Province Hospital of Integrated Chinese and Western Medicine were performed according to the Guide for the Care and Use of Laboratory Animals. A431 cells were subcutaneously injected into the right side of nude mice. When the tumor volume reached about 100 mm3, 5 mice were randomly selected and given a gavage of GSPs daily (200 mg/kg GSPs), and the other 5 mice were fed the distilled water as control group (denoted as 0 mg/kg GSPs). The length and width of the tumor were measured by vernier calipers to calculate the tumor volume (length × width2/2) every 4 days. After 28 days, the mice were sacrificed and the tumor was photographed and weighted. Tumor tissue was then collected for qRT-PCR and WB analysis.
Statistical Analysis
Data were expressed as mean ± standard deviation in three independent replicates. Statistical analyses were performed using GraphPad Prism 7 software (GraphPad Inc., La Jolla, CA, USA). Student’s t-test or one-way analysis of variance was used for comparing the differences between groups. P < 0.05 was defined as statistically significant.
Results
GSPs Reduced hsa_circ_0070934 Expression and Inhibited the Malignant Progression of CSCC
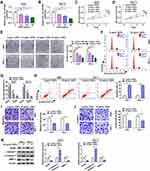
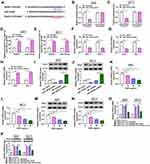
Under the treatment with different concentrations of GSPs, we found that the expression of hsa_circ_0070934 was significantly reduced in a concentration-dependent manner in A431 and SCL-1 cells (Figure 1A and B). To assess the effect of GSPs on the malignant progression of CSCC, we investigated CSCC cell progression under the treatment with different concentrations of GSPs. The results showed that as the increased concentration of GSPs, the viability and the number of colonies in A431 and SCL-1 cells were decreased significantly (Figure 1C–E). Subsequently, we assessed the cell cycle, apoptosis, migration and invasion ability in A431 and SCL-1 cells treated with 0 μg/mL and 20 μg/mL GSPs. Compared to control (0 μg/mL), GSPs (20 μg/mL) induced the cell cycle arrest in the G0/G1 phase and reduced the cell number in the S phase, as well as enhanced the apoptotic cells in A431 and SCL-1 cells (Figure 1F–H). In addition, the migrated and invaded A431 and SCL-1 cells were also remarkably suppressed under the treatment with GSPs (Figure 1I and J). More importantly, we noticed that the effects of different concentrations of GSPs on CSCC cell apoptosis, migration and invasion were also concentration-dependent (Supplementary Figure 1A–C). Moreover, GSPs also decreased the protein expression levels of proliferation marker PCNA and metastasis marker MMP-3, while increased apoptosis marker c-caspase 3/caspase 3 protein level in A431 and SCL-1 cells (Figure 1K). These data confirmed that GSPs could hinder CSCC cell progression.
Overexpression of hsa_circ_0070934 Reversed the Regulation of GSPs on CSCC Cell Progression
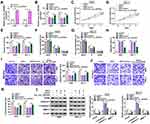
To identify the success of the amplified hsa_circ_0070934, we sequenced the PCR products of the hsa_circ_0070934 (Supplementary Figure 2A). Compared to GAPDH, hsa_circ_0070934 only could be amplified by divergent primers in cDNA but not in gDNA, suggesting that hsa_circ_0070934 had circular structure (Supplementary Figure 2B). In A431 and SCL-1 cells, we found that hsa_circ_0070934 was higher than that in HaCaT cells (Supplementary Figure 3A). To confirm whether GSPs regulated CSCC cell progression by regulating hsa_circ_0070934, hsa_circ_0070934 overexpression vector was constructed. After transfection of hsa_circ_0070934 overexpression vector into A431 and SCL-1 cells, the expression of hsa_circ_0070934 was significantly enhanced, confirming the transfection efficiency (Figure 2A). Then, A431 and SCL-1 cells were transfected with hsa_circ_0070934 overexpression vector or vector and then treated with 20 μg/mL GSPs. We found that the decreasing effect of GSPs on hsa_circ_0070934 expression could be abolished by the transfection of hsa_circ_0070934 overexpression vector (Figure 2B). The inhibitory effect of GSPs on the viability and the number of colonies and the promotion effect on the cell cycle arrest and apoptosis in A431 and SCL-1 cells could be reversed by overexpressing hsa_circ_0070934 (Figure 2C–H). Besides, overexpressed hsa_circ_0070934 also inverted the migrated and invaded A431 and SCL-1 cells inhibited by GSPs (Figure 2I–K). WB analysis results showed that hsa_circ_0070934 overexpression could reverse the suppressive effect of GSPs on the protein levels of PCNA and MMP-3 and the enhancing effect on c-caspase 3/caspase 3 protein level in A431 and SCL-1 cells (Figure 2L). All results revealed that GSPs inhibited CSCC progression by regulating hsa_circ_0070934.
Hsa_circ_0070934 Could Serve as a Sponge of miR-136-5p
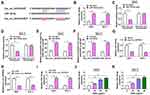
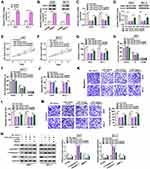
In order to elucidate the molecular mechanism by which GSPs regulated hsa_circ_0070934 and thus influencing CSCC progression, we used bioinformatics software to predict miRNAs that could interact with hsa_circ_0070934. The starBase software (http://starbase.sysu.edu.cn/starbase2/index.php) predicated that miR-136-5p had binding sites with hsa_circ_0070934 (Figure 3A). Subsequently, we constructed miR-136-5p mimic and confirmed that miR-136-5p mimic could indeed improve the expression of miR-136-5p in A431 and SCL-1 cells (Figure 3B). Then, miR-136-5p mimic or NC mimic and the pGL3 reporter vectors were co-transfected into A431 and SCL-1 cells to perform dual-luciferase reporter assay. The results showed that miR-136-5p mimic reduced the luciferase activity of pGL3-hsa_circ_0070934-WT vector, while had no effect on that of the pGL3-hsa_circ_0070934-MUT vector (Figure 3C and D). Furthermore, the enrichments of hsa_circ_0070934 and miR-136-5p could be increased in anti-AGO2 compared with that in anti-IgG (Figure 3E and F). MiR-136-5p expression could be significantly inhibited by overexpressing hsa_circ_0070934 in A431 and SCL-1 cells (Figure 3G). After silencing the hsa_circ_0070934 expression using si-hsa_circ_0070934, the expression of miR-136-5p was markedly increased in A431 and SCL-1 cells (Figure 3H and I). Compared to HaCaT cells, miR-136-5p was markedly downregulated in A431 and SCL-1 cells (Supplementary Figure 3B). In addition, miR-136-5p expression also was enhanced in A431 and SCL-1 cells with the increase in GSPs’ concentration (Figure 3J and K), which was contrary to the expression trend of hsa_circ_0070934. These results suggested that hsa_circ_0070934 could sponge miR-136-5p.
MiR-136-5p Reversed the Regulation of hsa_circ_0070934 on the Progression of GSPs-Treated CSCC Cells
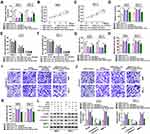
To confirm that GSPs regulated the hsa_circ_0070934/miR-136-5p axis to mediate CSCC progression, A431 and SCL-1 cells were co-transfected with hsa_circ_0070934 overexpression vector and miR-136-5p mimic, followed by treating with GSPs. The addition of miR-136-5p mimic promoted the miR-136-5p expression inhibited by hsa_circ_0070934 overexpression in GSPs-treated A431 and SCL-1 cells (Figure 4A). Then, we found that miR-136-5p mimic could reverse the enhancing effect of hsa_circ_0070934 overexpression on the viability, the number of colonies and cell cycle process, as well as the suppressing effect on the apoptosis in GSPs-treated A431 and SCL-1 cells (Figure 4B–G). Also, the migration and invasion of GSPs-treated A431 and SCL-1 cells promoted by hsa_circ_0070934 overexpression also could be abolished by miR-136-5p mimic (Figure 4H–K). In addition, miR-136-5p overexpression also reversed the promotion of hsa_circ_0070934 on PCNA and MMP-3 protein levels and the inhibition on c-caspase 3/caspase 3 protein level in GSPs-treated A431 and SCL-1 cells (Figure 4L). Therefore, all data confirmed that the hsa_circ_0070934/miR-136-5p axis was involved in the regulation of GSPs on CSCC progression.
MiR-136-5p Directly Targeted PRAF2
At the same time, the starBase software also was used to predict the targets of miR-136-5p and it was found that the 3ʹUTR of PRAF2 could bind to miR-136-5p (Figure 5A). Also, miR-136-5p mimic could inhibit the luciferase activity of pGL3-PRAF2 3ʹUTR-WT vector without affecting that of the pGL3-PRAF2 3ʹUTR-MUT vector (Figure 5B and C). Compared to anti-IgG, PRAF2 and miR-136-5p markedly enriched in anti-AGO2 (Figure 5D and E). Furthermore, we discovered that the mRNA expression of PRAF2 could be repressed by miR-136-5p mimic (Figure 5F), and it also could be promoted by miR-136-5p inhibitor after confirming the transfection efficiency of miR-136-5p inhibitor (Figure 5G–H). At the protein level, PRAF2 expression was also reduced by miR-136-5p mimic and enhanced by miR-136-5p inhibitor in A431 and SCL-1 cells (Figure 5I and J). In addition, we discovered that PRAF2 protein expression was also upregulated in A431 and SCL-1 cells compared to HaCaT cells (Supplementary Figure 3C). Moreover, under the treatment with different concentrations of GSPs, the mRNA and protein expression levels of PRAF2 were significantly decreased in a concentration-dependent manner (Figure 5K–N). Additionally, overexpressed hsa_circ_0070934 also could promote the mRNA and protein expression levels of PRAF2 in GSPs-treated A431 and SCL-1 cells, while these effects could be reversed by miR-136-5p mimic (Figure 5O and P). All results confirmed that PRAF2 was a target of miR-136-5p, and hsa_circ_0070934 could sponge miR-136-5p to regulate PRAF2.
MiR-136-5p Inhibited CSCC Cell Progression by Targeting PRAF2
The pcDNA PRAF2 overexpression vector was constructed and it was found that it could markedly enhance the mRNA and protein expression levels of PRAF2 in A431 and SCL-1 cells (Figure 6A and B). To further confirm that miR-136-5p indeed targeted PRAF2 to regulate CSCC progression, miR-136-5p mimic and pcDNA PRAF2 overexpression vector were co-transfected into A431 and SCL-1 cells. By detecting the mRNA and protein expression of PRAF2, we discovered that the decreasing of miR-136-5p mimic on PRAF2 expression could be recovered by pcDNA PRAF2 overexpression vector (Figure 6C and D). MiR-136-5p overexpression could suppress the cell viability and the number of colonies while inducing cell cycle arrest and apoptosis in A431 and SCL-1 cells. However, these effects could be reversed by PRAF2 overexpression (Figure 6E–J). Meanwhile, the numbers of migrated and invaded A431 and SCL-1 cells repressed by miR-136-5p mimic were inverted by overexpressing PRAF2 (Figure 6K–M). The inhibitory effect of miR-136-5p mimic on the protein levels of PCNA and MMP-3 and the promoting effect on c-caspase 3/caspase 3 also could be abolished by overexpressing PRAF2 (Figure 6N). These results demonstrated that miR-136-5p targeted PRAF2 to hinder CSCC progression.
GSPs Restrained CSCC Tumor Growth by Regulating the hsa_circ_0070934/miR-136-5p/PRAF2 Axis
For assessing the function of GSPs on CSCC tumor growth, we constructed a subcutaneous xenograft tumor using A431 cells. After GSPs treatment for 28 days, the tumor volume of the GSPs treatment group mice was markedly lower than the control group (Figure 7A). When the tumor was taken out for observation and weighting, we found that the tumor size and weight were significantly reduced in the GSPs treatment group (Figure 7B and C). To confirm the regulation of GSPs on CSCC tumor growth was mediated by the hsa_circ_0070934/miR-136-5p/PRAF2 axis, we detected the expression of hsa_circ_0070934, miR-136-5p and PRAF2 in the tumor tissues. The results showed that hsa_circ_0070934 expression was decreased, miR-136-5p expression was enhanced, and PRAF2 mRNA and protein expression was inhibited in the GSPs treatment group (Figure 7D–G). Hence, we confirmed that GSPs suppressed CSCC tumor growth via the hsa_circ_0070934/miR-136-5p/PRAF2 axis.
Discussion
In recent years, natural extracts have shown great potential in inhibiting the malignant progression of cancer. For example, resveratrol was found to suppress the invasion and metastasis of colon cancer,20 and curcumin had been discovered to have the potential to treat glioblastoma multiforme.21 However, the potential molecular mechanisms of natural extracts to exert anti-cancer effects remain to be further elucidated. Here, we investigated the effect of GSPs on CSCC progression. Compared to the previous study,11 we confirmed that GSPs had an inhibitory effect on CSCC cell invasion. In addition, we also showed that GSPs could hinder the proliferation, migration and cell cycle processes while promoting the apoptosis of CSCC cells. In animal experiments, mice given GSPs gavage showed smaller tumor volume and weight than the control group, which once again confirmed the anti-tumor effect of GSPs in vivo. These results provided the evidence that GSPs might have the potential to treat CSCC.
CircRNA is an important participant in human cancer progression and its high stability gives it great potential as a therapeutic target for cancer. In our study, we discovered that GSPs could decrease hsa_circ_0070934 expression in CSCC cells, and this effect was gradually enhanced with the increase of GSPs concentration. In past studies, hsa_circ_0070934 had been found to promote the proliferation and metastasis of CSCC.16,17 This suggested that hsa_circ_0070934 might play pro-cancer in CSCC. Here, our data showed that hsa_circ_0070934 overexpression could reverse the inhibitory effect of GSPs on CSCC progression, which confirmed that GSPs inhibited CSCC progression by decreasing hsa_circ_0070934 expression. Hsa_circ_0070934 also played an oncogenic role in CSCC in our study, which was similar to the previous results.16,17
Studies have confirmed that circRNA has many binding sites of miRNA, which can be used as the competitive endogenous RNA (ceRNA) to inhibit miRNA function.22 In order to reveal other new mechanisms by which hsa_circ_0070934 regulated CSCC progression, we used bioinformatics analysis to find that miR-136-5p could be targeted by hsa_circ_0070934. MiR-136-5p was lowly expressed in many cancers. Previous studies showed that miR-136-5p had anti-proliferation and anti-metastasis roles in cancers, such as renal cell carcinoma,23 cervical cancer24 and breast cancer.25 In addition, miR-136-5p also could suppress chondrocyte degeneration26 and was related to the inflammation of retinal pigment epithelial.27 Sand et al analyzed the differentially expressed miRNA in CSCC and normal skin tissue, and found that miR-136 was a significantly downregulated miRNA in CSCC.28 In this study, our data suggested that miR-136-5p could be promoted by GSPs in vitro and in vivo, and it also could reverse the enhancing effect of hsa_circ_0070934 on the progression of GSPs-treated CSCC cells. These results confirmed that GSPs reduced hsa_circ_0070934 expression to promote miR-136-5p expression, thereby regulating CSCC progression.
Further experiments revealed that PRAF2 was a target of miR-136-5p. PRAF2, a member of the PRA1 family, is an endosomal multi-pass membrane protein.29 Highly expressed PRAF2 can be detected in many cancers, and the high expression of PRAF2 often indicates poor prognosis for cancer patients.30,31 It had been reported that PRAF2 was overexpressed in CSCC and could promote CSCC migration and proliferation.32 In our study, the inhibitory effect of miR-136-5p on CSCC progression could be reversed by PRAF2, suggesting that PRAF2 also could facilitate CSCC proliferation, cell cycle process, migration, invasion, and reduce apoptosis. More importantly, the expression of PRAF2 could be decreased by GSPs and positively regulated by hsa_circ_0070934. These results confirmed the existence of the hsa_circ_0070934/miR-136-5p/PRAF2 axis and provided the evidence that GSPs regulated the hsa_circ_0070934/miR-136-5p/PRAF2 axis in CSCC.
Above all, our study revealed that GSPs suppressed CSCC progression, which was mainly achieved by regulating the hsa_circ_0070934/miR-136-5p/PRAF2 axis. Our findings illuminated the potential molecular mechanism of GSPs anti-cancer function, which provided a theoretical basis for GSPs to become an effective natural extract for CSCC treatment.
Highlights
1. GSPs inhibit CSCC cell growth and metastasis in vitro and tumor growth in vivo.
2. GSPs restrain CSCC progression by regulating hsa_circ_0070934.
3. Hsa_circ_0070934 serves as a sponge of miR-136-5p in CSCC.
4. MiR-136-5p targets PRAF2 in CSCC.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Waldman A, Schmults C. Cutaneous squamous cell carcinoma. Hematol Oncol Clin North Am. 2019;33(1):1–12. doi:10.1016/j.hoc.2018.08.001
2. Parekh V, Seykora JT. Cutaneous squamous cell carcinoma. Clin Lab Med. 2017;37(3):503–525. doi:10.1016/j.cll.2017.06.003
3. Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. 2018;78(2):237–247. doi:10.1016/j.jaad.2017.08.059
4. Green AC, Olsen CM. Cutaneous squamous cell carcinoma: an epidemiological review. Br J Dermatol. 2017;177(2):373–381. doi:10.1111/bjd.15324
5. Soura E, Gagari E, Stratigos A. Advanced cutaneous squamous cell carcinoma: how is it defined and what new therapeutic approaches are available? Curr Opin Oncol. 2019;31(5):461–468. doi:10.1097/CCO.0000000000000566
6. Burton KA, Ashack KA, Khachemoune A. Cutaneous squamous cell carcinoma: a review of high-risk and metastatic disease. Am J Clin Dermatol. 2016;17(5):491–508. doi:10.1007/s40257-016-0207-3
7. Pons Z, Guerrero L, Margalef M, et al. Effect of low molecular grape seed proanthocyanidins on blood pressure and lipid homeostasis in cafeteria diet-fed rats. J Physiol Biochem. 2014;70(2):629–637. doi:10.1007/s13105-014-0329-0
8. Gines I, Gil-Cardoso K, Terra X, et al. Grape seed proanthocyanidins target the enteroendocrine system in cafeteria-diet-fed rats. Mol Nutr Food Res. 2019;63(11):e1800912. doi:10.1002/mnfr.201800912
9. Wang W, Zhan L, Guo D, et al. Grape seed proanthocyanidins inhibit proliferation of pancreatic cancer cells by modulating microRNA expression. Oncol Lett. 2019;17(3):2777–2787. doi:10.3892/ol.2019.9887
10. Chen Q, Liu XF, Zheng PS, Yang BB. Grape seed proanthocyanidins (GSPs) inhibit the growth of cervical cancer by inducing apoptosis mediated by the mitochondrial pathway. PLoS One. 2014;9(9):e107045. doi:10.1371/journal.pone.0107045
11. Sun Q, Prasad R, Rosenthal E, et al. Grape seed proanthocyanidins inhibit the invasive potential of head and neck cutaneous squamous cell carcinoma cells by targeting EGFR expression and epithelial-to-mesenchymal transition. BMC Complement Altern Med. 2011;11:134. doi:10.1186/1472-6882-11-134
12. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
13. Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
14. Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234(8):14296–14305. doi:10.1002/jcp.28128
15. Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–352. doi:10.1016/j.bbrc.2018.09.073
16. An X, Liu X, Ma G, et al. Upregulated circular RNA circ_0070934 facilitates cutaneous squamous cell carcinoma cell growth and invasion by sponging miR-1238 and miR-1247-5p. Biochem Biophys Res Commun. 2019;513(2):380–385. doi:10.1016/j.bbrc.2019.04.017
17. Zhang DW, Wu HY, Zhu CR, et al. CircRNA hsa_circ_0070934 functions as a competitive endogenous RNA to regulate HOXB7 expression by sponging miR12363p in cutaneous squamous cell carcinoma. Int J Oncol. 2020;57(2):478–487. doi:10.3892/ijo.2020.5066
18. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
19. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
20. Yuan L, Zhou M, Huang D, et al. Resveratrol inhibits the invasion and metastasis of colon cancer through reversal of epithelial mesenchymal transition via the AKT/GSK3beta/Snail signaling pathway. Mol Med Rep. 2019;20(3):2783–2795. doi:10.3892/mmr.2019.10528
21. Shahcheraghi SH, Zangui M, Lotfi M, et al. Therapeutic potential of curcumin in the treatment of glioblastoma multiforme. Curr Pharm Des. 2019;25(3):333–342. doi:10.2174/1381612825666190313123704
22. Liu L, Wu SQ, Zhu X, et al. Analysis of ceRNA network identifies prognostic circRNA biomarkers in bladder cancer. Neoplasma. 2019;66(5):736–745. doi:10.4149/neo_2019_190107N25
23. Li J, Huang C, Zou Y, et al. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Mol Cancer. 2020;19(1):103. doi:10.1186/s12943-020-01225-2
24. Zhao J, Yang T, Li L. LncRNA FOXP4-AS1 is involved in cervical cancer progression via regulating miR-136-5p/CBX4 axis. Onco Targets Ther. 2020;13:2347–2355. doi:10.2147/OTT.S241818
25. Han C, Fu Y, Zeng N, et al. LncRNA FAM83H-AS1 promotes triple-negative breast cancer progression by regulating the miR-136-5p/metadherin axis. Aging (Albany NY). 2020;12(4):3594–3616. doi:10.18632/aging.102832
26. Chen X, Shi Y, Xue P, et al. Mesenchymal stem cell-derived exosomal microRNA-136-5p inhibits chondrocyte degeneration in traumatic osteoarthritis by targeting ELF3. Arthritis Res Ther. 2020;22(1):256. doi:10.1186/s13075-020-02325-6
27. Gao Z, Li Q, Zhang Y, et al. Ripasudil alleviated the inflammation of RPE cells by targeting the miR-136-5p/ROCK/NLRP3 pathway. BMC Ophthalmol. 2020;20(1):134. doi:10.1186/s12886-020-01400-5
28. Sand M, Skrygan M, Georgas D, et al. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2012;68(3):119–126. doi:10.1016/j.jdermsci.2012.09.004
29. Doly S, Marullo S. [PRAF2, an endoplasmic reticulum gatekeeper, controls the cell-surface export of the GABA(B) receptor in neurons]. Med Sci (Paris). 2015;31(10):834–836. French. doi:10.1051/medsci/20153110008
30. Wang CH, Liu LL, Liao DZ, et al. PRAF2 expression indicates unfavorable clinical outcome in hepatocellular carcinoma. Cancer Manag Res. 2018;10:2241–2248. doi:10.2147/CMAR.S166789
31. Qian Z, Wei B, Zhou Y, et al. PRAF2 overexpression predicts poor prognosis and promotes tumorigenesis in esophageal squamous cell carcinoma. BMC Cancer. 2019;19(1):585. doi:10.1186/s12885-019-5818-7
32. Yu GJ, Sun Y, Zhang DW, et al. Long non-coding RNA HOTAIR functions as a competitive endogenous RNA to regulate PRAF2 expression by sponging miR-326 in cutaneous squamous cell carcinoma. Cancer Cell Int. 2019;19:270. doi:10.1186/s12935-019-0992-x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.