Back to Journals » Infection and Drug Resistance » Volume 12
Genotypic characterization of Pseudomonas aeruginosa isolates from Turkish children with cystic fibrosis
Authors Sener Okur D , Yuruyen C , Gungor O, Aktas Z, Erturan Z, Akcakaya N, Camcioglu Y , Cokugras H, Koksalan K
Received 8 August 2018
Accepted for publication 15 February 2019
Published 27 March 2019 Volume 2019:12 Pages 675—685
DOI https://doi.org/10.2147/IDR.S183151
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Dicle Sener Okur,1 Caner Yuruyen,2 Ozge Gungor,2 Zerrin Aktas,2 Zayre Erturan,2 Necla Akcakaya,3 Yildiz Camcioglu,3 Haluk Cokugras,3 Kaya Koksalan4
1Istanbul University Cerrahpasa, Faculty of Medicine, Department of Pediatrics, Division of Pediatric Infectious Diseases, 34098 Kocamustafapasa, Istanbul, Turkey; 2Istanbul University Istanbul, Faculty of Medicine, Department of Microbiology and Clinical Microbiology, 34390 Capa, Istanbul, Turkey; 3Istanbul University Cerrahpasa, Faculty of Medicine, Department of Pediatrics, Division of Pediatric Infectious Diseases, Clinical Immunology and Allergy, 34098 Kocamustafapasa, Istanbul, Turkey; 4Istanbul University, Institute for Medical Experimental Research (DETAE), 34390 Capa, Istanbul, Turkey
Objective: To identify epidemic and other transmissible Pseudomonas aeruginosa strains, genotypic analyses are required. The aim of this study was to assess the distribution of P. aeruginosa strains within the Turkish pediatric cystic fibrosis (CF) clinic population.
Methods: Eighteen patients attending the pediatric CF clinic of Cerrahpasa Medical Faculty were investigated in the study. Throat swab and/or sputum samples were taken from each patient at 3-month intervals. The isolates of patients were analyzed by pulsed-field gel electrophoresis (PFGE). The intra- and interpatient genotypic heterogeneity of isolates was examined to determine the clonal isolates of P. aeruginosa within the cohort.
Results: A total of 108 clinical isolates of P. aeruginosa were obtained from 18 patients between May 2013 and May 2014. The pulsotypes of the first patient’s isolates could not be obtained by PFGE. From the remaining 17 patients and 101 isolates, 55 distinct pulsotypes were detected. The number of pulsotypes observed in more than one patient (minor clonal strains, cluster strains) was 8 (14.5%), and one of them colonized three patients. However, none of them was detected in more than three patients. These pulsotypes were composed of 20 isolates. In addition, with the PFGE analysis of 81 isolates, we detected 47 (85.6%) pulsotypes, which belonged to only one patient. Over different periods of this study, only 2 (11.8%) patients were colonized with the same pulsotype.
Conclusion: Our study indicates that there was considerable genomic diversity among the P. aeruginosa isolates in our clinic. The presence of shared pulsotypes supports cross-transmission between patients.
Keywords: P. aeruginosa, cystic fibrosis, PFGE, genotype, phenotype, epidemiology
Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative opportunistic pathogen in cystic fibrosis (CF) patients. It often causes chronic lung infection associated with respiratory failure, clinic deterioration, increased morbidity and mortality.1,2
 | Figure 1 Dendograms of the band profiles (first page). |
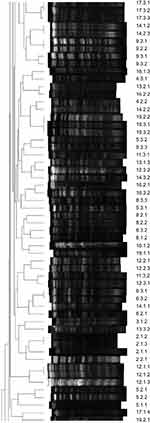 | Figure 2 Dendograms of the band profiles (second page). |
 | Figure 3 Dendograms of the band profiles (third page). |
It has been accepted that CF patients are colonized for long periods with their own unique P. aeruginosa strains acquired from the environment, and person-to-person transmission occurs infrequently.3–7 Recently, cross-infection and epidemic strains have been reported from the following different countries: United Kingdom, Australia, Norway, Germany, Canada and Denmark.8–15
In addition, the spread of an epidemic strain over several countries on two continents has been reported.16 Moreover, some epidemic strains are reported to be related to a poorer prognosis and the increased need for intravenous antibiotic treatment.10
To control and prevent P. aeruginosa infections among CF patients, it is necessary to determine if there is a persistence of the same strain or reinfection due to a new strain. Epidemic and other transmissible P. aeruginosa strains cannot be identified reliably by phenotypic markers and antibiotic susceptibility profiles. Therefore, genotypic analyses are required. Pulsed-field gel electrophoresis (PFGE) is the “gold standard” method for microbial typing.15,17–20
The epidemiological relatedness of P. aeruginosa isolates and the extent of cross-infection with these organisms among Turkish CF pediatric patients have not yet been investigated using PFGE. To our knowledge, this is the first published study on the genetic relatedness of P. aeruginosa isolates from CF pediatric patients in Turkey using the PFGE method.
The aim of this study was to assess the distribution of P. aeruginosa strains within the Turkish pediatric CF clinic population. The intra- and interpatient genotypic heterogeneity of isolates was examined to determine the clonal isolates of P. aeruginosa within the cohort.
Materials and methods
This study was approved by the Ethical Committee of Istanbul University Cerrahpasa Medical Faculty (date of approval 08/01/2013, approval number A-07) and conducted in accordance with the Declaration of Helsinki. Written informed consent to participate in the study was obtained from the patients enrolled or their parents.
Patients
Nineteen patients chronically infected with P. aeruginosa who attended the pediatric CF clinic of Cerrahpasa Medical Faculty were planned to be enrolled in the study. One patient left the study voluntarily, and the study was conducted with 18 patients. Their contact information was evaluated to explore possible routes of P. aeruginosa transmission among the infected patients. Half (female/male: 9/9) of our patients were female. The mean age of participants was calculated as 13±4.5 (1.83–22.08). Other clinical characteristics of patients’ are summarized in Table 1.
 | Table 1 Clinical profiles of patients’ |
Bacterial isolates
The isolates of P. aeruginosa from 18 children attending the pediatric CF clinic of Cerrahpasa Medical Faculty were collected over 1 year (May 2013 to May 2014). Throat swab and/or sputum samples were taken from each patient at 3-month intervals (May, August and November). The sputum samples were homogenized by mixing with dithiothreitol . The samples were inoculated onto MacConkey agar (Becton Dickinson, Franklin Lakes, NJ USA) and tryptic soy agar. Oxidase-positive Gram-negative bacilli colonies in different morphologies were further identified by the API 20 NE system (bioMérieux, Marcy-l'Étoile, France, Vitek Inc. ). The antibiotic susceptibilities of isolates were investigated in Müller–Hinton agar (Bio-Rad Laboratories Inc., Hercules, CA, USA) by the disc diffusion method according to the criteria of the Clinical and Laboratory Standards Institute (CLSI).21
The studied antibiotics were ofloxacin, ciprofloxacin, ceftazidime, imipenem, meropenem, ticarcillin-clavulanate, piperacillin, piperacillin-tazobactam, aztreonam, amikacin, gentamicin, netilmicin and tobramycin.
PFGE
PFGE was performed using a CHEF-DRIII drive module (Bio-Rad Laboratories Inc., Hercules, CA, USA). The isolates were grown overnight in tryptic soy agar at 37°C on a shaker. After standardization of the cell suspension by optical density measurement, the cells were embedded in low-melting agarose (Bio-Rad). Other steps, from the digestion (lysozyme and proteinase K) of bacteria to washing of the plugs and the subsequent restriction digestion of the bacterial DNA with SpeI (New England Biolabs, Ipswich, MA, USA) overnight, were performed in accordance with the kit manufacturer’s instructions. Electrophoresis was performed in 1% agarose gel prepared in 0.5x TBE (Tris-borate-EDTA) buffer. The running temperature was 14°C. The optimal run conditions for the separation of fragments were set as 18 hrs at 6 V/cm2 with an initial switch time of 6 s, a final switch time of 22 s and a 120° angle.
Lambda Ladder PFGE marker (BioLabs, Ipswich, MA, USA) was used as a molecular size marker. The gel was stained with ethidium bromide (1 µg/mL) for 30 mins, visualized under UV light using a transilluminator and photographed. A TIFF image of each gel was taken. The band profiles were analyzed by the GelCompar II system (version 6.0, Applied Maths, Sint-Martens-Latern, Belgium). Dendrograms of the band profiles were produced using the unweighted pair group method with mathematical averaging. The relatedness of isolates was calculated using the Dice coefficient with a band position tolerance setting of 1–1.5%. Isolates were defined as the same PFGE type (clonal) if the Dice coefficient was ≥85%. Tenover criteria were also applied for visual analysis of the bands22 (Figures 1–4).
 | Figure 4 Dendograms of the band profiles all in one page, with minor clonal strains coloured. |
Results
A total of 108 clinical isolates of P. aeruginosa were obtained from 18 patients between May 2013 and May 2014. According to the colonial morphology, the number of mucoid isolates (59%, 54.7%) was higher than the number of nonmucoid isolates (49%, 45.3%), and pigment production was observed in 21 (19.6%) isolates. The ratio of isolates sensitive to all antibiotics was 34.2% (37), and they were distributed among 15 patients (patients 1–4, 6, 8–15, 17 and 19). All the isolates of two patients (patients 10 and 19) were sensitive to all antibiotics. P. aeruginosa isolates were most susceptible to meropenem (98%, 90.7%), imipenem (96%, 88.8%), ciprofloxacin (95%, 87.9%) and piperacillin-tazobactam (95%, 87.9%). The numbers of mucoid isolates susceptible to all antibiotics (19%, 17.5) and nonmucoid isolates susceptible to all antibiotics (18%, 16.6%) were almost the same.
The pulsotypes of the first patient’s isolates could not be obtained by PFGE. From the remaining 17 patients and 101 isolates, 55 distinct pulsotypes were detected. The number of pulsotypes observed in more than one patient (minor clonal strains and cluster strains) was 8 (14.5%), and one of them colonized three patients. However, none of them was detected from more than three patients. These pulsotypes were composed of 20 isolates. From the remaining 81 isolates, 47 (85.6%) pulsotypes were detected, and each of them belonged to only one patient. (Table 2)
   | Table 2 Distribution of Pseudomonas aeruginosa isolates from 18 patients according to phenotypic feature and genotype (pulsotype) |
Only 2 (11.8%) patients (patients 2 and 15) were colonized with the same pulsotype (persistent pulsotype) over different periods of this study. Two (11.8%) patients (patients 14 and 17) harbored the same pulsotypes at the beginning of the study (the first two periods), and 5 (29.3%) patients (patients 6, 8, 9, 16 and 19) harbored the same pulsotypes in the last two periods. The other 8 (47.1%) patients’ (patients 3–5, 7 and 10–13) P. aeruginosa isolates all belonged to different pulsotypes in each study period. When the hospital records of these patients were examined, it was found that patient 15 had never been hospitalized and patient 2 had been hospitalized once in the past 5 years. Patients 6, 8 and 9 were hospitalized once in the study period between the second and third sample collection sessions. (Table 3)
 | Table 3 Genotypes of Pseudomonas aeruginosa isolates from 18 patients |
Our study indicates that there was considerable genomic diversity among the P. aeruginosa isolates in our clinic. The presence of shared pulsotypes supports cross-transmission between patients. In addition, we could not find a relationship between the genotype and phenotypic characteristics of P. aeruginosa isolates. Their antibiotic susceptibilities varied independently from their pulsotypes as well.
When we examined the patients’ contact information retrospectively from their hospital records, it was found that patient 10, a 13-month-old boy, had been hospitalized with patient 11 on the same dates several times. This finding was also observed in patients 3 and 4, as well as patients 12–14. However, there was no contact between patients 4 and 16, 4 and 14 or 16 and 19. Indeed, patients 16 and 19 were from different cities in Turkey. Infection prevention and control strategies in our CF center were carried out according to the 2003 North American guidelines.23 However, we experienced some difficulties fulfilling the recommendations. Health care personnel give great attention to hand hygiene, and although children and families are educated about respiratory hygiene practices, we observe that they sometimes do not comply with recommendations. Guidelines recommend that people with CF be placed in single-patient rooms, so we often hospitalize them in single-patient rooms. However, they must infrequently be placed in double-patient rooms due to our inpatient ward’s limited capacity. As a result, in our study, hospitalization was important for the transmission of strains. The transmission and acquisition routes of the pulsotype between patients 16 and 19 remain unclear. The source of this common pulsotype could be the environment. Furthermore, it could be a widespread strain. Inpatient rooms are cleaned and disinfected between patients, but unfortunately, this is not always possible for outpatient exam rooms. Generally, children with CF resist avoiding social and physical contact with other CF patients. Therefore, social or physical contact in outpatient waiting rooms may be another transmission route.
Discussion
This study explores the genotypic characterization of P. aeruginosa isolates collected from 18 patients at the pediatric CF center in Cerrahpasa Medical Faculty between 2013 and 2014. This is the first known published study exploring the genetic relatedness of P. aeruginosa isolates among CF children in Turkey. The identification of clonal, dominant or transmissible strains based on phenotypic features is not possible and instead requires molecular genotyping analyses.
One of the objectives was to establish if there was patient-to-patient transmission. Through PFGE analysis, we identified eight minor clones that included isolates from nine patients. The pulsotypes of the first patient’s isolates could not be obtained by PFGE. The endogenous endonucleases of the isolates could be the cause of this problem.
The presence of shared pulsotypes supports patient-to-patient transmission was possible. In the previous studies, which analyzed the PFGE patterns of P. aeruginosa isolates, the risk of transmission rates was extremely low.6,14,24,25
Similarly, the risk of transmission was low at our center. However, it cannot be ignored in some cases. It has been reported that different management of pulmonary exacerbations and infections due to P. aeruginosa is probably responsible for differences in epidemiology.26
Antimicrobial agents administered in the presence of pulmonary exacerbations, infection-control strategies in our center and efforts to eradicate P. aeruginosa might have prevented outbreaks of epidemic strains at our center. According to our infection-control program, for CF patients, we often hospitalized them in single-patient rooms. However, they must infrequently be placed in double-patient rooms due to our inpatient ward’s limited capacity. In this respect, our study is a guide for improving our infection-control program for children with CF. In addition, it is clear that we have to update our IPAC practices according to more recent guidelines published in 2013.27
The ability of P. aeruginosa to adapt and survive host immune responses, the extensive use of antibiotic therapy and the heterogeneity of the deteriorating lungs of CF patients cause clonal pathoadaptive variants with different phenotypic features.28
As this is the first known study that investigated the link between P. aeruginosa pulsotypes and phenotypic features, our phenotypic analysis showed that there is phenotypic heterogeneity within the pulsotype and between the minor clones as well. In addition, we observed that the isolates of shared pulsotypes did not have similar antimicrobial susceptibility patterns. Consistent with the previous studies, this study also found no relationship between the P. aeruginosa phenotype and genotype.29,30
Previous authors have reported that the majority of CF patients are colonized only with a unique genotype.14,30,31 However, coinfection with multiple P. aeruginosa isolates has also been observed in these patients.32–34
In our study, only two patients were determined to harbor just one P. aeruginosa genotype over the study period. Even though the design of the study was longitudinal, the results were different from those of previous studies, demonstrating that most patients were colonized for long periods with a single clone. More importantly, detecting several isolates with different genotypes in the same patient indicates that the colonizing strain may occasionally be replaced. Hospital admission and antimicrobial usage could also be responsible for these findings.
The main limitation of our study is the small sample size, but fewer children in our country are diagnosed with CF than in other countries. This may be because of differences in genetic predisposition between populations or the recent performance of newborn screening tests in our country. We are one of the main centers following children with CF in our country. More children with CF are followed by our center than most other CF centers in our country. Therefore, the number of patients we have followed is also low. In addition, we explored whether there is cross-transmission between patients by collecting samples over a year from each patient at 3-month intervals. We thought that taking three samples rather than one from each patient would increase the chance of detecting transmissible isolates.
One limitation of this study is that our investigation was a single-center study and the number of patients in our cohort was small. This study helped us recognize the need to organize a multicentre study. This study will also encourage other CF centers to investigate their P. aeruginosa isolates’ genetic relatedness and transmission properties and to collaborate with other researchers in this field.
Other two limitations of our study are that we did not perform environmental microbiological sampling, so we cannot comment on whether the transmissible pulsotypes were acquired from an environmental source and acquisition from social contact cannot be excluded.
Conclusions
This study suggested patient-to-patient transmission at our center by detection of shared clones (minor clones). To prevent the dissemination of this pathogen, infection-control strategies, such as the separation of patients and careful hygiene practices, should be followed.
Epidemiological and surveillance studies using molecular genotyping methods, although complex and expensive, help to monitor the emergence of epidemic clones and implement infection-control strategies within CF centers.
Further molecular epidemiological studies are needed to identify contaminated environmental sources and routes of patient-to-patient transmission and to improve infection-control and therapeutic measures. Longitudinal and multicentre studies are needed to explore the epidemic and widespread clones of other CF centers in Turkey.
Acknowledgments
This study was supported by the Istanbul University Scientific Research Projects Unit (IUBAP) (project number 29295).
Ethics approval and consent to participate
This study was approved by the Ethical Committee of Istanbul University Cerrahpasa Medical Faculty (date of approval 08/01/2013, approval number A-07) and conducted in accordance with the Declaration of Helsinki. Written informed consent to participate in the study was obtained from the patients enrolled or their parents.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Currie AJ, Speert DP, Davidson DJ. Pseudomonas aeruginosa: role in the pathogenesis of the CF lung lesion. Semin Respir Crit Care Med. 2003;24:671–680. doi:10.1055/s-2004-815663
2. Kosorok MR, Zeng L, West SE, et al. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol. 2001;32:277–287.
3. Boukadida J, De Montalembert M, Lenoir G, Scheinmann P, Veron M, Berche P. Molecular epidemiology of chronic pulmonary colonisation by Pseudomonas aeruginosa in cystic fibrosis. J Med Microbiol. 1993;38:29–33. doi:10.1099/00222615-38-1-29
4. Hunfeld KP, Schmidt C, Krackhardt B, et al. Risk of Pseudomonas aeruginosa cross-colonisation in patients with cystic fibrosis within a holiday camp-a molecular-epidemiological study. Wien Klin Wochenschr. 2000;112:329–333.
5. Pujana I, Gallego L, Martin G, Lopez F, Canduela J, Cisterna R. Epidemiological analysis of sequential Pseudomonas aeruginosa isolates from chronic bronchiectasis patients without cystic fibrosis. J Clin Microbiol. 1999;37:2071–2073.
6. Silbert S, Barth AL, Sader HS. Heterogeneity of Pseudomonas aeruginosa in Brazilian cystic fibrosis patients. J Clin Microbiol. 2001;39:3976–3981. doi:10.1128/JCM.39.11.3976-3981.2001
7. Struelens MJ, Schwam V, Deplano A, Baran D. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J Clin Microbiol. 1993;31:2320–2326.
8. Armstrong DS, Nixon GM, Carzino R, et al. Detection of a widespread clone of Pseudomonas aeruginosa in a pediatric cystic fibrosis clinic. Am J Respir Crit Care Med. 2002;166:983–987. doi:10.1164/rccm.200204-269OC
9. O’Carroll MR, Syrmis MW, Wainwright CE, et al. Clonal strains of Pseudomonas aeruginosa in paediatric and adult cystic fibrosis units. Eur Respir J. 2004;24:101–106.
10. Jones AM, Govan JR, Doherty CJ, et al. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet. 2001;358:557–558.
11. McCallum SJ, Corkill J, Gallagher M, Ledson MJ, Hart CA, Walshaw MJ. Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet. 2001;358:558–560.
12. Scott FW, Pitt TL. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J Med Microbiol. 2004;53:609–615. doi:10.1099/jmm.0.45620-0
13. Fluge G, Ojeniyi B, Hoiby N, et al. Typing of Pseudomonas aeruginosa strains in Norwegian cystic fibrosis patients. Clin Microbiol Infect. 2001;7:238–243.
14. Speert D, Campbell ME, Henry DA, et al. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am J Respir Crit Care Med. 2002;166:988–993.
15. Jelsbak L, Johansen HK, Frost AL, et al. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect Immun. 2007;75:2214–2224. doi:10.1128/IAI.01282-06
16. Römling U, Kader A, Sriramulu DD, Simm R, Kronvall G. Worldwide distribution of Pseudomonas aeruginosa clone C strains in the aquatic environment and cystic fibrosis patients. Environ Microbiol. 2005;7:1029–1038. doi:10.1111/j.1462-2920.2005.00780.x
17. Ruiz L, Domínguez MA, Ruiz N, Viñas M. Relationship between clinical and environmental isolates of Pseudomonas aeruginosa in a hospital setting. Arch Med Res. 2004;35:251–257. doi:10.1016/j.arcmed.2004.02.005
18. Johnson JK, Arduino SM, Stine OC, Johnson JA, Harris AD. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:3707–3712. doi:10.1128/JCM.00560-07
19. Waters V, Zlosnik JE, Yau YC, Speert DP, Aaron SD, Guttman DS. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis. 2012;31(12):3341–3350. doi:10.1007/s10096-012-1701-z
20. Wee BA, Tai AS, Sherrard LJ, et al. Whole genome sequencing reveals the emergence of a Pseudomonas aeruginosa shared strain sub-lineage among patients treated within a single cystic fibrosis centre. BMC Genomics. 2018;30(19):644. doi:10.1186/s12864-018-5018-x
21.
22. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239.
23. Saiman L, Siegel J,
24. Vosahlikova S, Drevinek P, Cinek O, et al. High genotypic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis in the Czech Republic. Res Microbiol. 2007;158:324–329. doi:10.1016/j.resmic.2007.02.003
25. Cigana C, Melotti P, Baldan R, et al. Genotypic and phenotypic relatedness of Pseudomonas aeruginosa isolates among the major cystic fibrosis patient cohort in Italy. BMC Microbiol. 2016;16:142. doi:10.1186/s12866-016-0760-1
26. Cramer N, Wiehlmann L, Ciofu O, Tamm S, Hoiby N, Tummler B. Molecular epidemiology of chronic Pseudomonas aeruginosa airway infections in cystic fibrosis. PLoS One. 2012;7:e50731. doi:10.1371/journal.pone.0050731
27. Saiman L, Siegel JD, LiPuma JJ, et al;
28. Bragonzi A, Paroni M, Nonis A, et al. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am J Respir Crit Care Med. 2009;180:138–145. doi:10.1164/rccm.200812-1943OC
29. Van Daele SG, Franckx H, Verhelst R, et al. Epidemiology of Pseudomonas aeruginosa in a cystic fibrosis rehabilitation centre. Eur Respir J. 2005;25:474–481. doi:10.1183/09031936.05.00050304
30. Leone I, Chirillo MG, Raso T, Zucca M, Savoia D. Phenotypic and genotypic characterization of Pseudomonas aeruginosa from cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 2008;27:1093–1099. doi:10.1007/s10096-008-0551-1
31. Logan C, Habington A, Lennon G, et al. Genetic relatedness of Pseudomonas aeruginosa isolates among a paediatric cystic fibrosis patient cohort in Ireland. J Med Microbiol. 2012;61:64–70. doi:10.1099/jmm.0.035642-0
32. Mowat E, Paterson S, Fothergill JL, et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med. 2011;183:1674–1679. doi:10.1164/rccm.201009-1430OC
33. Workentine ML, Sibley CD, Glezerson B, et al. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One. 2013;8:e60225. doi:10.1371/journal.pone.0060225
34. Finnan S, Morrissey JP, O’Gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol. 2004;42:5783–5792. doi:10.1128/JCM.42.12.5783-5792.2004
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
