Back to Journals » Infection and Drug Resistance » Volume 15
Genotypic and Phenotypic Expression of Antibiotic Resistance Patterns of Uropathogenic Enterobacteriaceae
Authors Gantasala E , Bhat S , Saralaya V, Jayaram M, Udayalaxmi J
Received 22 February 2022
Accepted for publication 17 May 2022
Published 26 July 2022 Volume 2022:15 Pages 3991—3999
DOI https://doi.org/10.2147/IDR.S362445
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Elizabeth Gantasala,1 Sevitha Bhat,2 Vishwas Saralaya,2 Madhumitha Jayaram,1 Jeppu Udayalaxmi2
1Kasturba Medical College, Mangalore, Manipal Academy of Higher Education, Manipal, Karnataka, India; 2Department of Microbiology, Kasturba Medical College, Mangalore, Manipal Academy of Higher Education, Manipal, Karnataka, 576104, India
Correspondence: Jeppu Udayalaxmi, Department of Microbiology, Kasturba Medical College, Mangalore, Manipal Academy of Higher Education, Manipal, Karnataka, 576104, India, Tel +91 824-2423452, Email [email protected]
Aim: To determine the antibiotic resistance patterns, detection of carbapenemase genes in uropathogenic bacilli belonging to the Enterobacteriaceae family and to correlate it with clinical data.
Materials and Methods: Identification and antibiotic sensitivity testing of the uropathogenic Enterobacteriaceae was done by using VITEK2 Compact (C) system. Multiplex PCR was used to detect blaIMP, blaKPC, blaNDM1, blaOXA − 48, and blaVIM genes.
Results: Out of 1602 urine samples, 417 (26%) showed significant growth, and in these 311 (74.6%) belonged to the Enterobacteriaceae family. Escherichia coli showed a relatively low rate of resistance to nitrofurantoin (17/205; 8.3%), with the majority of the isolates showing a MIC value of ≤ 16 μg/mL when compared to Klebsiella spp. (55/86; 64%), with MIC values for the majority of isolates being 128 μg/mL. Klebsiella spp. showed a relatively low rate of resistance to nalidixic acid (48/86; 55.8%) when compared with E. coli isolates (179/205; 87.3%). Out of 145 isolates tested, we found blaNDM in 11 (7.58%), bla OXA − 48 in 8 (5.51%), bla VIM in 4 (2.75%), bla KPC in one (0.6%) and blaIMP in none of the isolates. Of these 3 isolates were carbapenem sensitive, the rest were resistant.
Conclusion: Most of the isolates were sensitive to fosfomycin, carbapenems and resistant to cephalosporins and nalidixic acid. We detected carbapenemase genes in 13 (59%) out of 22 carbapenem resistant isolates and 3 (2.4%) out of 123 carbapenem sensitive isolates.
Keywords: antibacterial agents, drug resistance, multiplex polymerase chain reaction, urinary tract infection
Introduction
Enterobacteriaceae are normal flora of the human intestinal tract, but at times they can cause a number of diseases including urinary tract infections (UTIs), pneumonia, skin and soft tissue infections, and blood stream infections, in both community as well as hospital settings.1 UTIs caused by resistant strains of Enterobacteriaceae are a crucial public health problem, as they are associated with increased length of hospital stay, cost of treatment, morbidity and mortality.1,2 The resistance pattern of the uropathogenic bacteria varies with place and time, so periodic determination of antibiotic resistance patterns of these bacteria is required to decide upon the empirical therapy for a UTI.1,2
Beta-lactams which include penicillins, cephalosporins, monobactams and carbapenems are the most used antibiotics for treatment of Enterobacteriaceae infections. Their action is similar, as all of them inactivate the cell wall synthesis in bacteria.3,4 Carbapenems are last line drugs and are very effective against most of the beta-lactamases. Also they present very few side effects and are safer when compared to other last line drugs like polymyxin. But in recent times the Enterobacteriaceae are developing resistance to carbapenems.3,4 According to the Centers for Disease Control and Prevention (CDC) Enterobacteriaceae that are resistant to at least one of the carbapenem antibiotics namely ertapenem, doripenem, meropenem, or imipenem and/or possess carbapenemase gene are considered as carbapenem resistant Enterobacteriaceae (CRE).5 Carbapenem resistance in Enterobacteriaceae is mediated by overproduction of carbapenemase enzymes. Ambler classified all beta lactamases into four classes A-D. Klebsiella pneumoniae carbapenemase (KPC), New Delhi Metallo-beta-lactamase (NDM), Verona-Integron-Encoded Metallo-beta-lactamase (VIM), Imipenemase (IMP), and Oxacillinase −48 (OXA −48) are some of the carbapenemase enzymes which are most potent and prevalent among the Enterobacteriaceae.3,4 Ambler Class C enzymes (AMP C) have low potential for carbapenem hydrolysis, but may mediate efflux pump overexpression and/or reduced outer membrane permeability leading to carbapenem resistance in bacteria.3,4 In the present study the antibiotic sensitivity pattern of uropathogenic Enterobacteriaceae and the genes responsible for drug resistance namely blaIMP, blaKPC, blaNDM-1, blaOXA-48, and blaVIM were detected.
Materials and Methods
Study Design and Setting and Ethics Approval Statement
The study was conducted at the microbiology laboratory of a tertiary care center in South Karnataka. The study obtained approval from the Kasturba Medical College, Mangalore, Institutional Ethics Committee with approval no. IEC KMC MLR 11–19/594. Informed consent has been obtained from the patients for this study towards the use of samples and records. The study was performed as per the Declaration of Helsinki.
UTI cases which yielded >105 cfu/mL of pure growth of bacteria belonging to the Enterobacteriaceae family during the study period were included in the study. Urine samples which did not yield pure growth or significant growth were excluded.
Processing of Urine Samples
Urine samples were inoculated onto MacConkey’s agar, cystine lactose electrolyte deficient agar (CLED), and UTI chrome agar. Semi quantitative standard loop technique was used to determine colony count.6 The culture plates were incubated for 37 ℃ for 24 hrs. Urine samples which yielded pure growth of bacteria with a colony count of ≥105 CFU/mL were considered significant.6 The bacterial identification and antibiotic susceptibility testing (AST) was performed using Vitek Compact 2 system. (bioMerieux, North Carolina, USA). Vitek 2-GN card was used for identification and Vitek 2–AST–N235 card was used for antibiotic sensitivity with MIC values. Vitek Compact 2 system is an automated system which uses a microdilution technique as recommended by the CLSI7 also it uses the breakpoints recommended by the CLSI as well as EUCAST. Also Vitek 2 GN card contains an array of standard biochemical tests for the accurate identification of various gram negative bacilli. The clinical data of the patient was collected from the record section of the hospital.
The Detection of Resistance Genes
Carbapenem resistance genes were detected using a multiplex PCR assay. The DNA extraction was done using the boiling method. The primer sets were procured from Sigma Aldrich Pvt. Ltd (Table 1). Twenty microliter final volume multiplex PCR reaction mixture was prepared which contained, 2.6 µL master mix, 0.5 µL of each primer (10 x 0.5 µL; 5 µL), 10.4 µL nuclease-free water and 2 µL of extracted DNA. Amplification was done with cycling conditions; initial 5 min at 95 ℃ denaturation, 30 cycles of 30 secs at 95 ℃ denaturation, 60 secs at 60 ℃ annealing, 90 secs at 72 ℃ extension, followed by 10 min at 72 ℃ after the last cycle for the final extension. The amplified product was detected by agarose gel electrophoresis. The gel was then stained with 0.5 mg/mL ethidium bromide and visualized under UV transilluminator. Gel documentation system was used to obtain gel pictures (Alpha view 1.3.0, Alpha Innotech corporation MultiImage Light Cabinet)8–11 K. pneumoniae ATCC BAA 1705, K. pneumoniae BAA 1706, clinical isolates known to be positive for bla IMP, bla VIM, bla OXA 48, and bla NDM were used as controls.
 |
Table 1 Primers Used in the Multiplex PCR for Detection of Carbapenem Genes |
Statistical Analysis
Statistical analysis was performed using SPSS version 16.0. P value <0.05 was considered as significant.
Results
Out of 1602 urine samples that were received, 417 (26%) showed significant growth, among them, 311 (74.6%) belonged to the Enterobacteriaceae family. Out of these 311 patients, there were more women 203 (65.27%) than men 108 (34.7%) p < 0.05. A total of 168 (54.0%) samples were collected from adults (18–64 years), 125 (40.2%) belonged to the elderly population (≥65 years) and 18 (5.8%) were obtained from children (0–17 years). E. coli 205 (65.9%) was the most common organism, followed by Klebsiella spp. 86 (27.7%). The other Enterobacteriaceae found in the study were Enterobacter spp 6 (1.9%), Proteus spp. 6 (1.9%), Citrobacter spp. 4 (1.3%), Morganella spp. 2 (0.6%), Serratia spp. 1 (0.3%), and Providencia spp. 1 (0.3%).
Out of 160 ESBL producers 95 (59.3%) were isolated from female patients and 65 (40.6%) were male; out of 151 non ESBL producers, 108 (71.5%) were from female patients and 43 (28.47%) were from male patients. More numbers of male patients yielded ESBL producing organisms (p<0.05). There was no such correlation when compared with the age of the patient (p = 0.395).
Antibiotic Resistance Pattern of Uropathogens
The antibiotic sensitivity pattern of uropathogenic Enterobacteriaceae is shown in Table 2. The Enterobacteriaceae uropathogens under study were highly resistant to ampicillin (85.9%) with most of the isolates showing MIC values of ≥32 µg/mL, ticarcillin-clavulanate (79.7%) with most of the isolates showing MIC of ≥128 µg/mL and nalidixic acid (75.9%) with most of the isolates showing an MIC of ≥32 µg/mL. In comparison, they showed low resistance rates to piperacillin-tazobactam (14.1%), with most of the isolates showing an MIC of ≤4 µg/mL, ceftriaxone-sulbactam (14.1%), ertapenem (12.5%), with most of the isolates showing MIC of ≤5 µg/mL, meropenem (11.9%), amikacin (11.6%), with most of the isolates showing a MIC of ≤2 µg/mL, imipenem (8.7%), fosfomycin (4.5%), with most of the isolates having MIC of ≤16 µg/mL.
 |
Table 2 Antibiotic Sensitivity Pattern of Enterobacteriaceae Members Isolated from Cases of Urinary Tract Infection |
E. coli showed a relatively low rate of resistance to nitrofurantoin (17/205; 8.3%), with the majority of isolates showing an MIC value of ≤16 µg/mL and for all carbapenems tested, with MIC values of most of the isolates being <5 µg/mL. Most of the isolates of Klebsiella spp. were resistant to nitrofurantoin (55/86; 64%), with MIC values for the majority of isolates being 128 µg/mL. Klebsiella spp. showed a relatively low rate of resistance to nalidixic acid (48/86; 55.8%), with the MIC values ranging from <2 µg/mL to >32 µg/mL when compared with E. coli isolates (179/205; 87.3%) with most of the isolates showing an MIC value of >32 µg/mL (Figure 1).
 |
Figure 1 Comparison between antibiotic resistance pattern of E. coli and Klebsiella spp. |
Clinical Correlation
Out of 311 samples, complete clinical data was available for 139 cases (Figure 2). Most of the patients had multiple comorbidities, diabetes mellitus, hypertension, renal failure, carcinoma. Of the 139 cases, 91 (65.47%) were lower urinary tract infections and 48 (34.53%) cases were upper urinary tract infections. Among 48 cases of upper urinary tract infections there was one (2%) isolate of Proteus spp., 34 (24.46%) E. coli and 12 (25%) Klebsiella spp. Among 91 cases of lower urinary tract infection, 59 (64.8%) were E. coli, 24 (26.37%) Klebsiella spp., 3 (3.3%) Enterobacter spp., and 3 (3.3%) belonging to Tribe Proteeae one (1%) each of Citrobacter spp. and Serratia spp. There was not much difference in the etiology of upper and lower UTIs (p = 0.805).
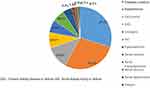 |
Figure 2 Coexisting clinical conditions in patients with UTI. Abbreviations: DM, diabetes mellitus; HTN, hypertension; CKD, chronic kidney disease or failure; AKI, acute kidney injury or failure. |
Molecular Characterization of Enterobacteriaceae Members Isolated from Cases of Urinary Tract Infection
Out of 311 Enterobacteriaceae isolates, 145 isolates could be preserved in pure form, which were used for carbapenemase gene detection. The rest of the isolates got discarded after the routine antibiotic sensitivity testing. A total of 24 genes were detected in 145 Enterobacteriaceae isolates, which included co-occurrence of two genes like blaOXA-48 and blaNDM; blaOXA-48 and blaVIM, and blaNDM and blaVIM. The distribution of the carbapenem genes among Enterobacteriaceae is summarized in Table 3. Figure 3 shows the gene detection done in the control strains.
 |
Table 3 Carbapenem Genes Detected Enterobacteriaceae Isolates from Cases of Urinary Tract Infection |
The most common gene identified was blaNDM, followed by both blaNDM and blaOXA48 together. Of 37 Klebsiella spp, 5 (13.5%) harbored blaNDM and 5 (13.5%) harbored blaOXA-48 and 3 (8.1%) harbored blaVIM genes. None of the Klebsiella spp. isolates carried blaKPC and blaIMP genes. However, 3 (8.1%) Klebsiella spp isolates co-harbored both blaNDM and blaOXA-48 genes; 1 (2.7%) blaOXA-48 and blaVIM; and 1 (2.7%) blaNDM and blaVIM genes. Out of 99 E. coli isolates, 6 (6.1%) harbored blaNDM, 3 (3%) blaOXA-48, 1 (1%) blaKPC, and 3 (3%) blaNDM and blaOXA-48. Out of 2 Morganella spp 1 (50%) harbored the blaVIM gene.
Out of 22 carbapenem resistant isolates we detected carbapenemase genes in 13 (59%) isolates, out of 123 carbapenem sensitive strains, we detected carbapenemase genes in 3 (2.4%) isolates. Greater numbers of carbapenem resistant isolates showed carbapenemase genes (p<0.001). Out of 79 ESBL producers we detected the carbapenemase genes in 14 (17.7%) isolates. Out of 66 non ESBL producers we detected carbapenemase genes in 2 (3%). Greater numbers of ESBL producers harbored the carbapenem genes (p = 0.005).
Discussion
Emerging problems of antibiotic resistance among uropathogens, requires frequent monitoring of their antibiotic susceptibility pattern to provide effective empirical therapy.1,2 In a study conducted in Jharkhand, India, UTI cases were greater among women than men, E. coli and Klebsiella spp. were common isolates. Almost half the isolates, 51.22%, were sensitive to nitrofurantoin in comparison to Klebsiella spp isolates, of which 25.8% were sensitive to nitrofurantoin. Most of the isolates of E. coli and Klebsiella spp. were resistant to nalidixic acid. Most of the E. coli isolates were sensitive to imipenem (90.8%) and meropenem (85.4%). Most of the Klebsiella spp. isolates were sensitive to imipenem (92%).12 A study conducted in Kerala showed a total of 373 positive urine samples with more patients being female (63.3%) than male (36.7%), E. coli (74.3%) was the most common isolate, followed by Klebsiella (15.8%). Cases were more in the age group 60–70. Both Klebsiella spp and E. coli were resistant to ampicillin and cephalosporins. E. coli isolates were 100% sensitive to piperacillin, amikacin, and imipenem followed by nitrofurantoin (90%) and cefoperazone (90%). Whereas 65% of Klebsiella spp. isolates were resistant to imipenem.13 In a study conducted in a children’s hospital in Pakistan, E. coli 211 (53.6%) and Klebsiella spp 157 (39.8%) were the predominant bacteria isolated. More than 84% of the bacterial strains were resistant to all of the cephalosporin drugs, 85% co-trimoxazole, 84.0% pipemidic acid, 83.5% nalidixic acid, 80.2% norfloxacin, 77.9% ciprofloxacin and 72.0% tobramycin. The isolates exhibited low resistance levels against amikacin (36.8%), fosfomycin (35.3%), nitrofurantoin (28.4%), imipenem (25.9%), and colistin (21.8%).14
The present study shows similar results, most of the UTI patients were women and E. coli was the predominant isolate. E. coli showed a relatively low rate of resistance to nitrofurantoin and for all carbapenems. Most of the isolates of Klebsiella spp. were resistant to nitrofurantoin and showed a relatively low rate of resistance to nalidixic acid when compared with E. coli isolates. In the present study 39% of uropathogenic Enterobacteriaceae showed carbapenem resistance. So generally carbapenem resistance in uropathogens vary from 8–65% in different geographical areas during different times.
In a study on bacteremia caused by coliforms, out of 68 isolates of gram negative bacilli resistant to carbapenems, 35 were AMP C producers, 25 were carbapenem resistant, 16 harbored blaNDM and none harbored bla KPC.9
In a study on carbapenem resistance in uropathogens, 66 carbapenem resistant isolates did not have any of the genes tested and nine isolates had multiple genes encoding for carbapenemase. None of the isolates from 2008 had blaNDM-1 or blaKPC genes, the blaVIM gene was present in 34 (43.6%) and blaIMP in five (6.4%). Among the isolates from 2012, none had blaKPC, but blaNDM-1 gene was present in 47 (53.4%), blaVIM in 19 (24.4%) and blaIMP in one (1.1%).10 In a study conducted in Iran, out of 211 clinical isolates of K. pneumoniae, 29 were carbapenem resistant strains. Of these 29 strains, 27 harbored the blaNDM-1 and 2 harbored the bla KPC gene.11
In a study conducted in Germany on 132 clinical Enterobacteriaceae strains with elevated carbapenem MICs, carbapenemase genes like blaKPC-2 (n = 13), blaKPC-3 (n = 11), blaKPC-2 and blaVIM-1 (n = 1), blaVIM-1 (n = 17), blaVIM-2 (n = 2), blaVIM-4 (n = 3), blaVIM-26 (n = 2), blaIMP-13 (n = 1), blaIMP-14 (n = 2), blaNDM-1 (n = 7), blaGIM-1 (n = 3), blaOXA-48 (n = 24), blaOXA-162 (n = 5), blaOXA-181 (n = 1), blaOXA-204 (n = 1), and blaGES-5 (n = 1) were detected by PCR and subsequent sequencing.15
In a study, 935 carbapenem resistant Enterobacteriaceae (CRE) strains were collected from 36 hospitals in different cities across China over a period of 2 years. Overall, carbapenemase genes were found in 97.4% of CRE strains. Among the genes detected were blaKPC-2 (51.6%), blaNDM (35.7%) and blaOXA-48 (7.3%). Overall, no strain was positive for blaVIM, the most prevalent carbapenemase gene was blaKPC-2 among K. pneumonia and blaNDM among E. coli. Most of the CRE strains were resistant to cephalosporins and more than half of the isolates were resistant to fluoroquinolones and aminoglycosides but the majority of the isolates were susceptible to tigecycline and polymyxin B.16
Phylogenetic studies have been extensively conducted on the uropathogenic E. coli (UPEC), as it is the predominant causative agent of urinary tract infections. UPEC has been classified into many phylogroups. Worldwide predominantly UPEC belong to phylogroups B2 and D. These phylogroups express most of the virulence factors and also are highly drug resistant.17 In a study, by pulse field gel electrophoresis (PFGE) UPEC belonged to 33 different genetic clusters and the majority belonged to PFGE type 11 which generated 15 bands.18 Horizontal gene transfer plays an important role in the spread of drug resistance. Integrons are genetic elements that are capable of integration of resistance gene cassettes. They integrate with transposons or conjugative plasmids and play a role in the spread of drug resistance.19 In a study, class 1 integron was detected in 52% of UPEC and these isolates were multi drug resistant.19 UTIs by bla CTX-M-15 gene harboring UPEC possess high virulence potential and high level of drug resistance.20 A study conducted in Iran showed that the UPEC isolates producing ESBL were more predominant in kidney transplant patients (KTP) when compared with a control group (non-KTP). UPEC bearing bla CTX-M and bla TEM were more prevalent in the KTP group when compared to the control group.21
In the present study, only carbapenemase genes were detected. The most common gene identified was blaNDM, followed by blaOXA-48, and blaNDM+OXA-48. Out of 22 carbapenem resistant isolates carbapenemase genes were detected in 13 (59%) isolates and out of 123 carbapenem sensitive strains, carbapenemase genes were detected in 3 (2.4%) isolates. We could collect clinical data from only 139 out of 311 cases, so not much clinical correlation could be done.
Conclusions
Out of 1602 suspected cases of urinary tract infection 417 (26%) showed significant growth, of which 311 (74.6%) urine samples yielded members of the Enterobacteriaceae family. Most of the cases of UTI were women. E. coli was the most common organism, followed by Klebsiella spp. Most of the isolates were resistant to ampicillin, ticarcillin clavulanate, and nalidixic acid and sensitive to piperacillin-tazobactam, ceftriaxone-sulbactam, ertapenem, meropenem, amikacin, imipenem, and fosfomycin. Carbapenem resistance was detected in 39% of the isolates. Carbapenemase genes were detected in 13 (59%) out of 22 carbapenem resistant isolates and 3 (2.4%) out of 123 carbapenem sensitive isolates. The most common gene identified was blaNDM, followed by both blaNDM and blaOXA48 together. None of the Klebsiella spp. isolates carried the blaKPC gene, and bla IMP gene was not detected in any of the isolates.
Acknowledgments
We are grateful to our institution for providing the infrastructure and other facilities to perform the study. We are grateful to Mrs. Swapna Bekal, English teacher and academic Co ordinator, Global City International School, Bangalore for doing the English grammar and flow editing. We are grateful to Dr. Dhanashree, associate professor, department of Microbiology, KMC, Mangalore for her inputs regarding the methodology and final draft of the article.
Disclosure
The authors declare that they have no conflicts of interest in relation to this work.
References
1. Wang JT, Wu UI, Lauderdale TLY, et al. Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS One. 2015;10(3):1–18.
2. Pollett S, Miller S, Hindler J, Uslan D, Carvalho M, Humphries RM. Phenotypic and molecular characteristics of carbapenem-resistant Enterobacteriaceae in a health care system in Los Angeles, California, from 2011 to 2013. J Clin Microbiol. 2014;52(11):4003–4009. doi:10.1128/JCM.01397-14
3. Sahin K, Tekin A, Ozdas S, et al. Evaluation of carbapenem resistance using phenotypic and genotypic techniques in Enterobacteriaceae isolates. Ann Clin Microbiol Antimicrob. 2015;14(1):1–6. doi:10.1186/s12941-015-0105-1
4. Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):1521. doi:10.1177/2049936115621709
5. Centers for Disease Control and Prevention. Definition of Carbapenem resistant Enterobacteriaceae. Available from: https://www.cdc.gov/hai/organisms/cre/technical-info.html.
6. Collee JG, Duguid JP, Fraser AG, Marmion BP, Simmons A. Laboratory strategy in the diagnosis of infective syndromes. In: Collee JG, Fraser AG, Marmion BP, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology.
7. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
8. Pragasam AK, Sahni RD, Anandan S, et al. A pilot study on Carbapenemase detection: do we see the same level of agreement as with the CLSI observations. J Clin Diagnostic Res. 2016;10(7):DC09–DC13.
9. Porwal A, Bhat S, Hegde A, Rao P, Shenoy S. Clinicomicrobiological study of bacteraemia caused by coliforms in adults. J Clin Diagnostic Res. 2018;12(7):DC15–9.
10. Mohan B, Hallur V, Singh G, Sandhu HK, Appannanavar SB, Taneja N. Occurrence of bla NDM-1 & absence of bla KPC genes encoding carbapenem resistance in uropathogens from a tertiary care centre from north India. Indian J Med Res. 2015;142(3):336–343. doi:10.4103/0971-5916.166601
11. Hosseinzadeh Z, Sedigh Ebrahim-Saraie H, Sarvari J, et al. Emerge of bla NDM-1 and bla OXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J Chin Med Assoc. 2018;81:536–540. doi:10.1016/j.jcma.2017.08.015
12. Kumar A, Kumar R, Gari M, Keshri USP, Mahato SK, Ranjeeta K. Antimicrobial susceptibility pattern of urine culture isolates in a tertiary care hospital of Jharkhand, India. Int J Basic Clin Pharmacol. 2017;6(7):1733–1739. doi:10.18203/2319-2003.ijbcp20172740
13. Jubina Bency AT, Priyanka R, Jose P. A study on the bacteriological profile of urinary tract infection in adults and their antibiotic sensitivity pattern in a tertiary care hospital in central Kerala, India. Int J Res Med Sci. 2017;5(2):666–669. doi:10.18203/2320-6012.ijrms20170171
14. Ejaz H, Ahsan A, Zafar A, et al. Bacterial profile and antimicrobial resistance of uropathogenic Enterobacteriaceae. Pakistan J Med Heal Sci. 2019;13(2):451–454.
15. Martin K, Florian S, Lars W, Gatermanna G. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J Clin Microbiol. 2012;50(9):3115–3118. doi:10.1128/JCM.00991-12
16. Han R, Shi Q, Wu S, et al.; and the China Antimicrobial Surveillance Network (CHINET) Study Group (2020). Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
17. Halaji M, Fayyazi A, Rajabnia M, Zare D, Pournajaf A, Ranjbar R. Phylogenetic group distribution of uropathogenic Escherichia coli and related antimicrobial resistance pattern: a meta-: analysis and systematic review. Front Cell Infect Microbiol. 2022;12:790184. doi:10.3389/fcimb.2022.790184
18. Raeispour M, Ranjbar R. Antibiotic resistance, virulence factors and genotyping of uropathogenic Escherichia coli strains. Antimicrob Resist Infect Control. 2018;7(1):118. doi:10.1186/s13756-018-0411-4
19. Khoramrooz SS, Sharifi A, Yazdanpanah M, et al. High frequency of Class 1 Integrons in Escherichia coli isolated from patients with urinary tract infections in Yasuj, Iran. Iran Red Crescent Med J. 2016;18(1):e26399. doi:10.5812/ircmj.26399
20. Ogbolu DO, Terry Alli OA, Webber MA, Oluremi AS, Oloyede OM. CTX-M-15 is established in most multidrug-resistant uropathogenic Enterobacteriaceae and Pseudomonaceae from hospitals in Nigeria. Eur J Microbiol Immunol. 2018;8(1):20–24. doi:10.1556/1886.2017.00012
21. Halaji M, Shahidi S, Atapour A, Ataei B, Feizi A, Havaei SA. Characterization of Extended-Spectrum β-Lactamase producing uropathogenic Escherichia coli among Iranian Kidney Transplant Patients. Infect Drug Resist. 2020;13:1429–1437. doi:10.2147/IDR.S248572
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.