Back to Journals » Infection and Drug Resistance » Volume 12
Genotype Analysis of Clinical Candida albicans Isolates Using PCRs Targeting 25S rDNA and ALT Repeat Sequences of the RPS and Antifungal Susceptibility in Ouagadougou (Burkina Faso)
Authors Sawadogo PM, Zida A, Soulama I , Sermé SS, Guiguemdé KT , Junior R, Sangaré I , Bamba S, Ouédraogo-Traoré R, Guiguemdé TR
Received 4 August 2019
Accepted for publication 1 December 2019
Published 16 December 2019 Volume 2019:12 Pages 3859—3866
DOI https://doi.org/10.2147/IDR.S225947
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eric Nulens
Patindoilba Marcel Sawadogo,1–3 Adama Zida,1–3 Issiaka Soulama,3,4 S Samuel Sermé,4 Kiswendsida Thierry Guiguemdé,2,5 Richard Junior,1 Ibrahim Sangaré,5,6 Sanata Bamba,5,6 Rasmata Ouédraogo-Traoré,2,7 Tinga Robert Guiguemdé6,8
1Parasitology-Mycology Department, University Hospital Yalgado Ouédraogo, Ouagadougou, Burkina Faso; 2Training and Research Unit in Health Sciences, University Joseph Ki-Zerbo (UO JKZ), Ouagadougou, Burkina Faso; 3Private School of Health News Sciences (ESSN), Ouagadougou, Burkina Faso; 4National Center for Research and Training on Malaria, Ouagadougou, Burkina Faso; 5Parasitology-Mycology Department, Souro Sanou University Hospital, Bobo Dioulasso, Burkina Faso; 6High Institute of Health Sciences, University Nazi Boni, Bobo Dioulasso, Burkina Faso; 7Laboratory Department, University Hospital Charles De Gaulles, Ouagadougou, Burkina Faso; 8Muraz Research Center, Bobo-Dioulasso, Burkina Faso
Correspondence: Patindoilba Marcel Sawadogo
Parasitology-Mycology Department, University Hospital Yalgado Ouédraogo, 03 BP 7022 Ouaga 03, Ouagadougou, Burkina Faso
Tel +22672112786
Email [email protected]
Objective: Candida albicans is a yeast with multiple genotypes. It’s a commensal fungus colonizing various sites. However, when the host’s immune system weakens, it becomes pathogenic and is responsible for various lesions. In Burkina Faso, antifungal drugs are frequently used, particularly fluconazole, the most used systemic antifungal. This antifungal drug and other antifungal drugs are often used for self-medication or prescribed outside of antifungal susceptibility test results. These situations led to the emergence of Candida albicans strains resistant to antifungal drugs commonly used in Burkina Faso. The aim of this study was to determine the types of Candida albicans using PCRs targeting 25S rDNA and ALT repeat sequences of the RPS and to establish their azoles and polyenes susceptibility profile.
Material and methods: Antifungal susceptibility testing by disk diffusion method was performed in accordance with CLSI document M44-A for yeasts and the manufacturer’s instructions. Candida albicans isolates were genotyped using specific PCR primers of the rDNA and RPS genes.
Results: Ten (10) RPS types of Candida albicans were found in our study: The most common RPS types are A3 (40.6%), A2 (24.0%) and A2/3 (14.6%) for genotype A, B2/3 (5.2%) for genotype B and C2 (3.2%) for genotype C. The Azole resistance, especially fluconazole (74.4%), was the most common with genotype A, including A3 (36.6%), A2 (18. 3%). Polyene resistance was rare with nystatin, only A3 (1.2%) resistant isolate to nystatin was observed. For amphotericin B, the highest observed resistance rates were A3 (11.0%) and A2/3 (8.5%) for the genotype A and B2 (10.0%), B3 (10.0%) and B2/3 (10.0%) for genotype B.
Conclusion: Our study showed that Candida albicans resistance to azoles, especially to fluconazole, is an important phenomenon in Ouagadougou, and several genotypes RPS types are involved. Thus, fluconazole would not be an antifungal agent for first-line prescribing for treatment of candidiasis in Ouagadougou. This study will be continued to determine the molecular mechanisms involved in these antifungal resistances, for further research of new molecules with different action targets.
Keywords: Candida, rDNA, RPS, antifungal, susceptibility, Burkina Faso
Introduction
Candida albicans is a commensal fungus, colonizing various sites such as the vagina, mouth and gastrointestinal tract.2 However, when the host’s immune system weakens, it becomes pathogenic and is responsible for various lesions. In the majority of cases, there are superficial mucosal lesions, but it can enter the bloodstream and cause disseminated candidiasis.3
Several genotypes of C. albicans are involved in candidiasis.18 PCR targeting 25SrDNA, which has been frequently used for genotype analysis of C. albicans, allows C. albicans to be grouped into five genotypes: A, B, C, D, and E.6 In addition, it has been demonstrated that genotype A is the most common genotype in different regions.2 A study conducted in 2016 in Burkina Faso, showed that C albicans genotypes found in Ouagadougou are A, B and C.18 Genotype A was present in all the sources of isolation such as vulvovaginal swabs, Nails, Broncho-alveolar lavage fluid, whereas genotypes B and C are frequently found in oral sampling.18
In Burkina Faso, antifungal drugs are frequently used, particularly fluconazole, the most used systemic antifungal. This antifungal drug and other antifungal drugs are often used for self-medication or prescribed outside of antifungal susceptibility test results. These situations led to the emergence of C. albicans resistant strains to antifungal drugs commonly used in Burkina Faso; and all three circulating genotypes are involved in resistance to antifungal drugs, especially to azoles.18 For example, fluconazole used for candidiasis preventive treatment for people living with Human immuno-deficiency virus (HIV) in Burkina Faso, is 65% less effective against C. albicans.18 Fluconazole is also the most accessible antifungal drug to the public. Other azoles are often neither available nor accessible. To prevent this risk, it is imperative to look for new molecules with different mechanisms of action. For that, we need to know the different mechanisms of C albicans resistance to antifungal drug in the local context.
According to the literature, mechanisms of C. albicans resistance to antifungal drugs are diverse and depend in part on the family of antifungals, the strain and sometimes the locality.13 The most common mechanism of C. albicans resistance to azoles involves the ERG11 gene specially mutations on this gene. Mutations have also been demonstrated on other genes conferring resistance to other molecules such as CDR1 and 2, MRR1 and 2, TAC1 and others ERG genes for polyenes, allylamines and azoles, FKS1 for echinocandins, FCA1 and FCY1 for pyrimidine analogues.13
The aim of this study was to determine the types of C. albicans using PCRs targeting 25S rDNA and ALT repeat sequences of the RPS and to establish their azoles and polyenes susceptibility profile.
Materials and Methods
Isolates Used
Ninety-six (96) C. albicans clinical isolates from various sites (oral cavity, vagina, nails, smooth skin, urine and stools) were used. A C. albicans reference strain ATCC10231 (Laboratoire Humeau) was used as a PCR positive control.
Antifungal Susceptibility Testing
Antifungal susceptibility testing by disk diffusion method was performed in accordance with CLSI document M44-A for yeasts and the manufacturer’s instructions. C. albicans isolates were subcultured on Sabouraud Dextrose Agar. Saline suspensions of isolates were made and the turbidity was adjusted at 0.5 McFarland standards. A lawn culture was done on freshly prepared Muller Hinton Agar plates. Antifungal disks were placed onto the surface of each of the inoculated agar plate and read after 24 h and 48 h. The antifungal disks tested were azoles as fluconazole (100 μg), itraconazole (50 μg), econazole (10 μg), clotrimazole (50 μg), ketoconazole (10 μg), miconazole (10 μg), associated with polyenes including amphotericin B (20 μg) and nystatin (100 IU), all produced by Liofilchem R _ s.r.l. (Italy). The isolates were classified as susceptible, susceptible dose-dependent or resistant according to the CLSI guidelines.18
Extraction of DNA from Isolates
The extraction of the DNA was done in two successive stages. A first step consisted of enzymatic lysis with betamercaptoethanol and lyticase (Sigma Aldrich, ≠ M3148). It allows to obtain spheroplasts. The second step consisted in the extraction of the DNA itself by the kit NucleoSpin® Tissue kit (Macherey-Nagel).18
Primers Used for PCR
For genotype determination, the primers used were based on 25S rDNA including direct primers CA-INT-L: ATA AGG GAA GTC GGC AAA ATA GAT CCG TAA and reverse primer CA-INT-R: CCT TGG CTG TGG TTT CGC (Table 1).10
 |
Table 1 List of PCR Primers (25SrDNA and RPS) and Expected Sizes of PCR Products |
For the determination of the different subtypes based on ALT (tandem short repeating unit of 172b) Repetitive Characteristic Sequences (RPS), the primer pair (ASDcF: TGA ACC TGA ACT TGT GCT ACA AAG and pCSCR: CGC CTC TAT TGG AGC TCG AGT AGT C) (Gen Bank. accession nos. L47111) was used. These primers can divide the C. albicans isolates into 6 subtypes depending on the size of the PCR products (Table 1).2 All primers were provided by Sigma Aldrich.
Conditions for PCR Amplification and Agarose Gel Electrophoresis
DNA was amplified in a reaction mixture (25 μL) containing 1.75 mM MgCl 2, 0.2 mM dNTPmix, 1 U Taq DNA polymerase (Fermentase), 50 pmol d primers and 2 μL of DNA. The condition of the PCR was as follows: incubation at 95°C for 3 mins followed by 35 cycles at 94°C for 30 seconds, 59°C for 30 seconds, 72°C for 30 seconds and 72°C for 10 mins. The PCR products were electrophoresed on a 1.2% agarose gel and stained with ethidium bromide for 20 mins at 20°C.2 PCR master mix is also provided by Sigma Aldrich
Ethical Considerations
For the collection of strains, Ethical approval was obtained from the national ethic commission of Burkina Faso. Written informed consent was obtained from all study participants, after explaining to them the importance of the study
Results
25S rDNA Based Genotyping
All C. albicans isolates were identified by two specific pairs of oligonucleotide primers (Gen Bank accession nos. L47111, L28817) (Figure 1). The genotypes of all understudy isolates of C. albicans were analyzed by the PCR method. PCR amplification of 450, 840, and both 450 and 840 bp DNA products which was performed using 25S rDNA, corresponded to genotypes A, B and C, respectively (Figure 1)
Results presented in Table 2 revealed that among 96 isolates, 82 were classified as genotype A (85,4%), 10 were classified as genotype B (10,4%) and 4 were classified as genotype C (4,2%). In addition, genotypes D and E were not found in our study
 |
Table 2 Frequency and Distribution of Candida albicans Genotypes According to Sources of Isolation |
Genotype A was predominant at 76.3% in oral sampling. Moreover, it was found in all vaginal sampling. Genotypes B and C were exclusively isolated from oral sampling (Table 2).
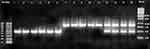 |
Figure 1 Amplification profiles of PCR products (25S rDNA) of Candida albicans isolates. |
The molecular size marker (100 bp) is in the line marked M and the corresponding sizes, the base pairs being given on the left and on the right. Pathways (1–6) were genotype A (approximately 450 bp), pathways (7–11) were genotype B (approximately 840 bp), and pathways (12–15) were genotype C (approximately 450 and 840 bp). bp).
RPS Based Genotyping
The RPS-based genotyping of the isolates was determined on the basis of the repeated numbers of the ALT sequence in the major product that showed the highest intensity (Figure 2). As illustrated in Table 3, genotypes A, B and C of C. albicans were classified into genotype RPS types (Figure 2). The 82 A genotypes were classified into A2 (24.0%), A3 (40.6%), A2/3 (14.6%) and A3/4 (6.3%). The 10 B genotypes were considered to be B2 (2.1%), B3 (2.1%), B2/3 (5.2%) and B3/4 (1.0%). The 4 C genotypes were grouped into C2 (3.1%) and C2/3 (1.0%) genotypes (Table 3).
 |
Figure 2 Variations in Candida albicans RPS types based on PCR amplification profiles using RPS primers. |
 |
Table 3 Analysis of Candida albicans RPS Types by RPS Primers |
C. albicans has been classified into 4 RPS types based on the major band number with the highest intensity. The molecular size marker (1 kb) was in the first and last lines marked M, and the corresponding sizes in base pairs were given on the left and right. Lanes (1–3) were of RPS type 2 (698 bp), lanes (5–6) were of RPS type 3 (870 bp), lanes (4, 7–9) were of RPS type 2/3 (698 and 870 bp) and lanes 10 and 11 were of RPS 3/4 (870 and 1042 bp).
Antifungals Susceptibility
Azoles Susceptibility
Resistance to azole is present with all three genotypes (Table 4). It is more common with genotype A strains (all azoles, excepted econazole), than with strains of the other two genotypes B and C. The highest resistance of genotype A strains were found with fluconazole, itraconazole and ketoconazole: 74.4%; 68.4% and 25.6% respectively. The differences are not statistically significant (p˃0.05) (Table 4).
 |
Table 4 Azoles and Polyenes Susceptibility Profiles of Candida albicans Genotypes and RPS Types |
A3 (36.6%) and A2 (18.3%) RPS types were the most resistant to fluconazole. Itraconazole is the second azole most affected by the resistance of our strains. We noted that A3 at 30.5% and A2 at 20.8% were also the most resistant to itraconazole. Ketoconazole is the third azole that has experienced more resistance to our strains, especially with the RPS type A3 (12.2%) and A2 (7.3%). The resistance of our strains with miconazole and clotrimazole was rarely observed. Only the RPS type A3 is involved in the resistance to these two antifungal drugs at respectively 1.2% and 2.4%. We did not observe genotype A resistance to econazole (Table 4).
Regarding genotype B, we noted the following observations: B3 (20.0%), B 2/3 (30.0%) were the only resistant RPS types to fluconazole. RPS type B2/3 is also largely involved in itraconazole resistance with 40.0% of all B genotypes. Genotype B resistance to ketoconazole was rarely observed. It was absent with miconazole, econazole and clotrimazole (Table 4).
Two RPS types of genotype C, especially C2 (50.0%) were resistant to fluconazole. All C2 (75.0%) were resistant to itraconazole and the only RPS type C2/3 was resistant to ketoconazole (Table 4).
Polyenes Susceptibility
Only one RPS type of genotype A, especially A3 (1.2%) was resistant to nystatin. Regarding amphotericin B, the highest resistance rate was observed with the RPS types A3 (11.0%) and A2/3 (8.5%) (Table 4).
We did not observe nystatin-resistant strains with the B genotype. The B2 (10.0%), B3 (10.0%) and B2/3 (10.0%) RPS types were resistant to amphotericin B (Table 4). No strain of genotype C was resistant to polyene (Table 4).
Discussion
PCR targeting 25S rDNA confirmed that genotype A (85.4%) accounted for the majority of isolates, followed by genotype B (10.4%) and C (4.2%). Genotypes D and E were not found in our results. In Japan authors14 analyzed 301 isolates of C. albicans and classified them into 4 genotypes; A (132 isolates), B (66 isolates), C (56 isolates) and D (5 isolates). Other authors reported that genotype A had more frequency in invasive and non-invasive isolates of C. albicans.5,7 In opposition to our results, previous studies have found that genotypes A and C were more common in non-invasive and invasive isolates, respectively.3,4 Such genotyping based on 25S rDNA was largely adapted for C albicans and the genotype A of C. albicans constituted the majority of isolates in all previous reports.10,12,16 In a study,6 the ratio of C. albicans genotype B or C to genotype A varied in each group of clinical samples. These findings may be affected by the types of clinical specimens colonized by C. albicans, the geographic location and the different patient populations over time.
Each genotype A, B and C has been grouped into at least two RPS types. The majority of genotype A, B and C RPS types were A3 (40.6%), B2/3 (5.2%) and C2 (3.2%), respectively. In general, A3 constitutes the majority of clinical isolates. These results are similar to those found in another study, in which C. albicans was isolated from various sites, vaginal secretions, sputum and blood and subjected to RPS-based genotyping.6 Azole resistance was found to be more frequent with genotype A. The highest resistance rates with genotype A were obtained with fluconazole (74.4%) followed by itraconazole (68.4%) (Table 2). RPS types A3, B2/3 and C2 are the RPS types of each genotype, the most involved in the resistance [Table 3]. According to the literature, of all commonly used antifungal families, the resistance of C. albicans to azoles, especially fluconazole, is the most common.15 Our results are in agreement with the conclusions of a study conducted in Nigeria where genotype A with its RPS type A3 showed high resistance to fluconazole.5 The RPS types, most implicated in azole resistance including A3, would be more susceptible to mutations conferring them this resistance.15 Thus, according previous data, the RPS types most involved in azole resistance would have more mutation on ERG genes including the ERG11 gene.15
The presence of transposable group I intron in 25S rDNA decreases the level of resistance to fluconazole and itraconazole and largely to azoles. However, in another context where only vaginal isolates are the only ones involved in resistance, the presence of group I introns transposable at this level has been found to be associated with an increased level of resistance to fluconazole and itraconazole.17
In addition it has also been recognized that chromosomes of C. albicans contain characteristic repetitive sequences (RPS) defined as ALT which represents the nucleotide sequences of the internal repeats of the RPS.11 The number of ALT repeats in the RPS varies in each chromosome, thus leading to a variation in the molecular sizes of the RPS. These molecular characteristics are exploited for the typing of C. albicans.2
Resistance to nystatin was rare. We observed only one genotype A isolate (1.2%) including the resistant A3. This phenomenon of resistance to nystatin was absent with genotypes B and C (Table 3). A similar observation was also noted in a previous study.8 For the second polyene, amphotericin B, we found relatively high levels of resistance (30% and 29.3%). The presence or absence of group 1 intron in 25S rDNA from a less intron (genotype A with A3) and an intron (genotype B, with equal B2, B3 and B2/3) are involved (Table 2). It has also observed an absence of association between the presence of the group 1 intron in C. albicans 25S rDNA and its susceptibility to amphotericin B.1 The resistance of C. albicans to this polyene has been increasingly reported in recent years from several clinical isolates.17 However lower resistance rates to amphotericin B have been reported in other studies. For example, in a study involving children with neutropenia4 and another9 with samples from patients with genital tract infection reported resistance however to amphotericin B in lower rates of 4% and 0.5%, respectively.
Conclusion
Genotyping of C albicans from 25S rDNA in our study showed the presence of three A, B and C genotypes. These three genotypes were classified into 10 RPS types following genotyping by the ALT repeats in the RPS sequences. This study will be continued to determine the molecular mechanisms involved in these antifungal resistances, for further research of new molecules with different action targets.
Acknowledgments
This work was supported by the Department of Parasitology-Mycology of the University Hospital Yalgado Ouédraogo. Our acknowledgments to: Mr. Constant Dahourou, General mananger of University Hospital Yalgado Ouédraogo and all his staff; Dr. Adama Gansané, Managing Director of National Center for Research and Training on Malaria (CNRFP); all patients who participated in this study.
Disclosure
This work and an abstract of this document was presented at the 8th Congress of the West African Society of Parasitology (SOAP) in Bamako (Mali) in December 2016 in the form of an oral presentation presenting provisional partial results. The abstract of the presentation was published in “Presentation Abstracts” in the journal Medical Mycology (2017, 0, 1-4)/Oxford Academic: DOI: 10.1093/mmy/myx127. The authors report no other conflicts of interest in this work.
References
1. MS A-S, Jamous RM, Alothman NHA, et al. Genotyping and antifungal susceptibility of Candida albicans strains from patients with vulvovaginal and cutaneous candidiasis in Palestine. Afr J Microbiol Res. 2015;9:952–959.
2. Ashrafi Iradj T, Zahraei Taghi S, Sharifzadeh A, Shokri H, Khosravi AR. Repetitive sequences based on genotyping of Candida albicans isolates obtained from Iranian patients with human immunodeficiency virus. Iran J Basic Med Sci. 2014;17:831–835.
3. Gurbuz M, Kaleli I. Molecular analysis of Candida albicans isolates from clinical specimens. Mycopathologia. 2010;169:261–267.
4. Haddadi P, Zareifar S, Badiee P, et al. Yeast colonization and drug susceptibility pattern in the pediatric patients with neutropenia. Jundishapur J Microbiol. 2014;7:11858.
5. Hattori H, Iwata T, Nakagawa Y, et al. Genotype analysis of Candida albicans isolates obtained from different body location of patients with superficial candidiasis using PCRs targeting 25S rDNA and ALT repeat sequences of the RPS. J Dermatol Sci. 2017;42:31–46.
6. Iwata T, Hattori H, Chibana H, et al. Genotyping of Candida albicans on the basis of polymorphisms of ALT repeats in the repetitive sequence (RPS). J Dermatol Sci. 2006;41:43–54.
7. Karahan ZC, Guriz H, Agrbasal H, et al. Genotype distribution of Candida albicans isolates by 25S intron analysis with regard to invasiveness. Mycosis. 2014;47:465–469.
8. Li JY, Sun HY, Zhang QQ. Antifungal susceptibility test of genotypes of Candida albicans from patients with atrophic or erosive oral lichen planus. Shanghai Kou Qiang Yi Xue. 2011;20:300–303.
9. Luo X, Dong X, Distribution PZ. Drug susceptibility of Candida spp. associated with female genital tract infection, Chongqing, China. Jundishapur J Microbiol. 2015;8(9):e19386.
10. McCullough MJ, Clemons KV, Stevens DA. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J Clin Microbiol. 1999;37:417–421.
11. Mijiti J, Pu XM, Erfan A, Yaguchi T, Chibana H, Tanaka R. Genotyping of fluconazole-resistant Candida albicans isolated from Uighurian people in Xinjing (China) using ALTS/RFLP and micro-TGGE method. Nihon Ishinkin Gakkai Zasshi. 2010;51:165–168.
12. Millar BC, Moore JE, Xu J, Walker MJ, Hedderwick S, McMullan R. Genotypic subgrouping of clinical isolates of Candida albicans and Candida dubliniensis by 25S intron analysis. Lett Appl Microbiol. 2002;35:102–106.
13. Sawadogo PM, Zida A, Sangaré I, et al. Genetic mutations conferring resistance to candida albicans to antifungal drugs: a global perspective and regional implications. J Infectiology. 2019;2(4):6–12.
14. Tamura M, Watanabe K, Mikami Y, Yazawa K, Nishimura K. Molecular characterization of new clinical isolates of Candida albicans and C. dubliniensis in Japan: analysis reveals a new genotype of C. albicans with group I intron. J Clin Microbiol. 2016;39:4309–4315.
15. Tapia CV, Hermosilla G, Fortes P, et al. Genotyping and persistence of candida albicans from pregnant women with vulvovaginal candidiasis. Mycopathologia. 2017;182:339–347.
16. Vrioni G, Matsiota-Bernard P. Molecular typing of Candida isolates from patients hospitalized in an intensive care unit. J Infect. 2011;42:50–56.
17. Zarei Mahmoudabadi A, Zarrin M, Kiasat N. Biofilm formation and susceptibility to amphotericin B and fluconazole in Candida albicans. Jundishapur J Microbiol. 2014;7:28–36.
18. Zida A, Soulama I, Bamba S, et al. 25S rDNA genotype and antifungal susceptibility of clinical Candida albicans in Ouagadougou (Burkina Faso). Med Mycol. 2017;56:61–65.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
