Back to Journals » International Journal of General Medicine » Volume 13
Genomics and Transcriptomics: The Powerful Technologies in Precision Medicine
Authors Khodadadian A , Darzi S, Haghi-Daredeh S , Sadat Eshaghi F, Babakhanzadeh E, Mirabutalebi SH, Nazari M
Received 15 February 2020
Accepted for publication 4 September 2020
Published 17 September 2020 Volume 2020:13 Pages 627—640
DOI https://doi.org/10.2147/IJGM.S249970
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Ali Khodadadian,1 Somaye Darzi,1 Saeed Haghi-Daredeh,2 Farzaneh Sadat Eshaghi,3 Emad Babakhanzadeh,1,4 Seyed Hamidreza Mirabutalebi,1 Majid Nazari1
1Department of Medical Genetics, Faculty of Medicine, Shahid Sadoughi University of Medical Sciences, Yazd, Iran; 2Department of Medical Nanotechnology, School of Medicine, Shahroud University of Medical Sciences, Shahroud, Iran; 3Department of Medical Genetics, Biotechnology Research Center, International Campus, Shahid Sadoughi University of Science, Yazd, Iran; 4Yazd Medical Genetics Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran
Correspondence: Ali Khodadadian Email [email protected]
Abstract: In a clinical trial, people with the same disease can show different responses after treatment with the same drug and exactly under the same conditions. Some of them may improve, some may not show any response, and occasionally side effects may be observed. In other words, people with the same disease process under the same therapeutic conditions may have different responses. Today, some diseases are resistant to conventional (standard) treatment procedures. Why do people with the same disease show different responses to the treatment with the same drug? This is primarily due to differences in molecular pathways (especially genetic variations) associated with the disease. On the other hand, designing and delivery of a new drug is a time-consuming and costly process, so any mistake in any stage of this process can have irreparable consequences for pharmaceutical companies and consumer patients. Therefore, we can achieve more accurate and reliable treatments by acquiring precise insight into different aspects of precision medicine including genomics and transcriptomics. The aim of this paper is to address the role of genomics and transcriptomics in precision medicine.
Keywords: precision medicine, personalized medicine, genomics, transcriptomics, pharmacology
Background
Currently, many diseases are not predictable in the early-stage, and there is no definitive treatment for some of them (eg, neurodegenerative diseases and most cancers).1,2 In addition, many available medical treatments are designed for a wide range of patients with the same condition that, after prescribing medication, the following states may be observed: 1) The prescribed drug may reduce the symptoms of the disease and improve the treatment process;3,4 2) The expected response may fail to occur,5,6 and 3) The prescribed medication can cause side effects in patients.4,7 We must note that all of these conditions may be observed in people who suffered from the same disease and receive a similar medication. Different responses in the same treatment are often due to genetic, environmental, and lifestyle differences between patients.8 In other words, according to precision cancer medicine, each person is unique, and so is his or her cancer; this concept can be generalized to other conditions as well. For example, in cystic fibrosis and diabetes (or any similar condition) different forms of the disease can be caused by various mutations or factors (multifactorial etiologies).9,10 Then, each of these forms may require unique therapeutic procedures to achieve effective treatment and reduce the symptoms of the disease. In order to achieve strategies for overcoming these problems and finding more precise treatments, scientists and researchers have always tried to provide the best treatments according to their knowledge of the nature of the disease and individual characteristics of patients. The history of these activities dates back to many years ago when our ancestors tried to the healing of some diseases by tailoring treatment regimens based on patient’s characteristics while they did not know much about the nature of these diseases. Alternatively, as an example of more precise treatment methods in recent years, improvements in the blood donation process can be noted; in which the donor selection is based on the existence of the same characteristics between the donor and recipient.11,12 Recent developments in the field of genetics, particularly the identification of disease-related variations in the genome of patients through identifying single nucleotide polymorphism (SNP) genotyping (via executing some studies such as the Human Genome Project and Genome Wide Association Studies) has led to a dramatic increase in our understanding of the causes of diseases.13 These improvements led to the advent of a new field in diagnosing and treating diseases as well as reducing healthcare costs.14–16 This new field is called precision medicine (Figure 1). A simple definition for precision medicine is a treatment designed for diseases that, unlike the conventional procedures, do not prescribe a common treatment for all people with the same disease. Rather, it divides patients into subgroups based on their common characteristics, and then prescribes a specific treatment for each subgroup; therefore, minimizing the cost and side effects as well as maximizing the therapeutic efficacy will be achieved. The distinctive features of patients can be genotypic profiles, clinical manifestations, and lifestyle. In general, it includes anything causing heterogeneity in the treatment of the same illness under the same therapeutic procedures.17–20 Taken together, precision medicine involves understanding the diseases and characteristics of patients at deeper levels, and then designing drugs and therapies according to these findings. A well-known example of the application of modern precision medicine is designing of the ivacaftor and lumacaftor for cystic fibrosis patients.21 These drugs are designed for two subgroups of CF patients who have a different mutation in CFTR gene.22–26 Another example is BiDil. BiDil was certified in 2005 for treating congestive heart failure, but only in African-Americans.27
Precision Medicine
Precision medicine is also known as personalized medicine and individualized medicine. The precision medicine concept was perceived by our ancestors centuries ago when they prescribed special dietary supplements to the patients based on their experiences. Another clear example is related to Jews who had refused circumcision for the sons of all sisters of a mother who had sons with the “bleeding disease” (hemophilia) 2000 years ago.28 The term “personalized medicine” is older than precision medicine, but nowadays precision medicine is preferred (Figure 2). One reason for this renaming is that the term “personalized medicine” may lead to misunderstanding. For instance, it may be thought that the purpose of personalized medicine is the designing of a unique treatment for each patient, while this interpretation is not exactly right. The more correct and practical meaning of this term is tailoring treatment for a subgroup of patients who have similar characteristics that differentiate them from other subgroups in the same disease.21,29,30 In addition, the P4 term (predictive, preventive, personalized, and participatory) medicine is also used in some cases to describe this particular form of medicine31 and these terms are still used interchangeably.32 However, we use the precision medicine term in the current article; and this article aimed to review the following: the requirements and prerequisites for precision medicine, the most important advances and achievements in precision medicine as well as and some of the diseases whose treatment has been influenced by precision medicine were discussed.
Hypothesis
Nowadays the time of trial and error to achieve the desired outcomes has been passed; especially, if the target society consisting of human patients. Accordingly, before ensuring the results of a new trial, drug, procedure, etc. by testing on experimental models and passing the initial stages, it cannot be introduced into the human world. These steps may take years and are staggeringly costly.33,34 Therefore, it is very important to detect any trouble in the earliest phase in this process before too much time and funds have been spent. In this regard, determining genotype and predicting potential related side effects are very important and any negligence could cause irreparable consequences for patients and drug companies. For example, an anti-inflammatory drug, Rofecoxib, was withdrawn in 2004 from the market only five years after its public release, since it was proven that this drug can lead to increased risk of heart attack and stroke in some people who took high doses of it.35 Thus, identification of potential effects of a new drug before its presentation to the market is very important. However, an important issue which should not be ignored is that a particular drug may result in different responses in people with the same disease (especially based on their genetic characteristics). For example, in the case of Rofecoxib, it can be said that this drug only causes side effects in people with a specific genotype, and it not leading to side effects in other people with other genotypes in the same condition. In this case, maybe there was no need to remove the drug from the market and suppliers could continue selling the drug via a genetic test, and the patients would benefit from the drug too. Altogether, precision medicine can play an important role in the realization of this goal (ie, designing and prescription of more precise drugs). Investment in this field can help us to achieve better treatment procedure in some of the diseases, particularly in relation to diseases in which there are no (at least by now) definitive cures. In this respect at least we will encounter two important problems: i) the cost of designing new drugs is so high that all people cannot use it; and ii) pharmaceutical companies tend to design drugs that are utilizable for a wide range of patients. Both of these defects are contradicting with the main goals of precision medicine. In the study, we will review some of the most important aspects of precision medicine as well as its pitfalls.
Requirements
The emergence and entry of omics studies has changed and improved our understanding of human pathophysiology and encouraged the progress of the precision medicine field.36 There are 4 key elements required for a full understanding of precision medicine. 1) Genomics, to get a deeper understanding of disease by discovering specific genetic changes (in the genome of the patients) which have an association with the disease.37 2) Transcriptomics, a technique that obtains information on the abundance of multiple mRNA transcripts within a biological sample simultaneously. 3) Proteomics, completing and confirming the information obtained through genomics by studying of all of the proteins.38,39 4) Metabolomics, investigating the cellular metabolites from biological processes to obtain a physiological snapshot of the cell.40 In addition to the above, there are other elements that are important which should not be ignored, including a) providing the tools and databases to collect, integrate, and share the obtained information of the research in order to accelerate the improvements in precision medicine; b) translating information obtained from research data into clinical applications (Figure 3).
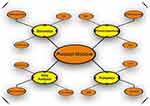 |
Figure 3 Most important fields as well as some of their components and achievements affecting precision medicine. |
Genomics
After the discovery of the DNA structure by Francis Harry Compton Crick and James Dewey Watson in 1953,41 a huge revolution took place in all branches of biological sciences. Among these changes, the following milestones can be mentioned: a) the discovery of the first sequencing methods by Sanger and Coulson42,43 and Maxam and Gilbert;44 b) the discovery of the polymerase chain reaction in 1985 by Kary B. Mullis.45 A few years later, these achievements provided the basic elements for precision medicine. Most of the initial activities for understanding and developing precision medicine have focused on genomics as the source of potential differences related to disease across populations.
What is the genome? The term “genome”, as a set of the haploid chromosomes, was first used in the “Spread and cause of pathogenesis in plant and animal kingdoms” book, written by German botanist Hans Winkler in 1920.46 Today, the genome refers to any genetic material in any type of organism. The genome includes chromosomal and extrachromosomal DNA material, either coding (contains coding and non-coding genes which may or may not have a protein product) and non-coding region.47–50 Note that a distinction should be made between non-coding genes and the non-coding region of the genome. Although the former is non-coding, it is a region which contains genes that are transcripted (to functional RNA) but not translated, while the latter is a region without any gene and often includes repetitive sequences that are not transcripted. Genomics is an interdisciplinary field of science that studies the entire genome. Unlike genetics, which examines the individual characteristics of genes, genomics aims to: identify the interaction between the genes and the synergistic effects of them, the interaction of environment and genes, as well as the study of the non-coding regions of the genome.51,52 Genomic medicine is an interdisciplinary medical area including the use of genomic information that has promptly developed since the completion of the Human Genome Project (HGP) more than a decade ago and it includes concepts such as Biomarker, Codon, DAN, Exome, Exon, and so on.53
The completion of the Human Genome Project (HGR) in 2003 has led to a dramatic increase in our understanding of the role of genome in molecular mechanisms of disease.54–56 Also, the knowledge gained through further studies in the field of genomics has led to changes and contributed to the incorporation of genetics into clinical medicine.57 Two key elements in genome studies are sequencing and data analysis, each one requiring its own tools and expertise. With the possession of these skills and expertise, after discovering and analyzing the genetic differences associated with a condition in patients, it is possible to design a treatment for individuals based on their own genetic and environmental characteristics in order to reduce costs and side effects. It is not surprising if we consider the human genome project as one of the first milestones in the modern precision medicine. On the other hand, entry of the next-generation sequencing (NGS) methods into the genome-sequencing field has created a massive development in the precision medicine applications. NGS can be considered another milestone as another milestone in the avenue of development of the precision medicine.
Why is human genome sequencing important? There are many reasons for the importance of sequencing the genome, but the most important of them are the following. a) DNA underlies approximately every feature of human health, in both normal and abnormal situations; therefore, by having DNA sequences in both normal and abnormal forms we can find effective differences between patients. b) It is crucial to study gene expression in a specific tissue, organ, or tumor.58 c) For studying human variation,36 it is necessary to understand how genome alterations are related to the development of cancer. d) It is highly useful in determining the rate of sensitivity to the drug in the same disease for patients, based on their DNA sequences (pharmacogenomics).36 Genome sequencing can help us to discover the precise molecular pathways associated with the disease; it is a vital element in precision medicine.
Transcriptomics
The transcriptome refers to all of the RNA transcripts in a cell or tissue.59 Transcriptome includes alternative splice variants.60,61 One of the main reasons for the study of the transcriptome is that most human genes undergo a process called alternative splicing. As it is known, a large number of eukaryotic genes are composed of exons and introns. On the other hand, only exons are translated into protein products, while introns are removed after transcription. The process via which introns removed from an mRNA and exons are reconnected again is called splicing (Figure 4).
 |
Figure 4 Splicing: the process by which the introns are removed and the exons are re-connected. |
Alternative splicing is a particular type of splicing in which two or more rearrangements (removal of introns and reconnection of exons) can occur in one mRNA in separate tissues or different stages of cell (tissue) growth. Therefore, several proteins can be produced from one mRNA without any changes in its related gene (Figure 5). For example, through the alternative splicing, the nuclear pre-RNAs from fibronectin gene are converted to 20 mRNAs, encoding 20 different proteins.62 Thus, from the fibronectin gene, 20 mRNAs and 20 proteins are produced without any alteration in the fibronectin gene (without any alteration in genomics level). Alternative splicing affects most activities of human genes (95% of multi-exonic genes).63,64 The occurrence of errors at this level can lead to many diseases such as β+-thalassaemia (a disorder characterized by reduced β-globin protein levels and anaemia), Duchenne muscular dystrophy (mutations in splice site of dystrophin gene and loss of dystrophin protein function) and other examples.64 Accordingly, any disturbance in this pathway can lead to disease. Note that these defects are not detectable at the genomics level.
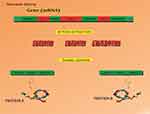 |
Figure 5 Alternative splicing is a particular type of splicing in which, after the removal of the introns, the exons are joined in different ways. |
Transcriptomics is the study of transcriptome and is involved in the function of cells, tissues, or organisms, across a wide range of biological conditions. The relationship between the transcriptome and the genome suggests that the information of an organism is stored in the DNA (the genome) and expressed by RNA (the transcriptome). The main focus of the transcriptomics is to discover how transcripts of a cell, tissue, or living organism are influenced by disease or environmental factors (such as drugs, hormones, etc.). Non-coding RNA is another very important aspect of transcriptomics. These functional elements play an important role in the occurrence of a variety of diseases and their response to treatment.65–67
The transcriptomics includes the post-transcriptional era; hence, the alterations that cannot be detected at the genomics level can be revealed in the transcriptomics level. Then, if these changes have pathogenic importance, they should be identified in order to design more reliable treatments. In other words, the transcriptomics is a continuum and complements the genomics and fills the gap between genomics and proteomics in the precision medicine era. There are different methods for transcriptome analysis, depending on whether the examination is to be performed on a single gene (or a small number of genes) or all transcripts of a cell, tissue or organism. Some of the most important of these methods are listed in Table 1.
 |
Table 1 Most Common Methods for Transcriptome Study |
In recent years, genomics and transcriptomics have been widely used in clinical applications. Some of the most recent of these studies are presented in Table 2.
 |
Table 2 Some of the Most Recent Clinical Applications of Precision Medicine |
Well-Known Examples of Applications of Precision Medicine
Today, there are many examples regarding clinical applications of precision medicine and its effects on diseases. Some of the examples of clinical applications of precision medicine are mentioned in different sections of this article. Herein, in order to gain a comprehensive insight into precision medicine applications, we are going to describe well-detailed examples.
Pharmacogenetics is perhaps the first precision medicine application68 and is defined as a study of genetic differences and its effect on drug metabolism pathways, which ultimately lead to different responses to the same drug in different individuals according to their genetic characteristics.69,70 One of the first examples of the pharmacogenetics is the sum of the trials performed for genotyping VKORC1 and CYP2C9 to adjust warfarin dosing. Finally, these trials led to some success such as methods for automated dose estimation in patients who received warfarin.68 Compared with pharmacogenetics, a term widely used for genes which are involved in drug metabolism directly, the pharmacogenomics term covers all genomic interactions involved in drug metabolism.71 Accordingly, if we want to summarize the main goal of pharmacogenetics or pharmacogenomics, it tries to determine the genotype of a patient and prescribe the most appropriate dosage of the best drug for her or him according to her or his genotype which has an association with the disease (ie, genotypes that influence the function of the drug). Therefore, pharmacogenetics or pharmacogenomics can be considered as one of the precision medicine achievements.
Cystic Fibrosis
One of the best-known examples of application of the precision medicine and its role in improving disease treatment is its role in improving cystic fibrosis (CF) patients. CF is an autosomal recessive disorder caused by mutations in the cystic fibrosis transmembrane conductance (CFTR) gene (located on 7q31.2). CF was first recognized as a distinct disease in 1936 and was known as ‘mucoviscidosis’ because of the deposit of thick mucus secretions leading to obstruction of the airways and secondary infection.28 In 1987, the location of the gene responsible for the CF disease was detected on q-arm of chromosome 7.72 Finally, in 1989 this gene was identified, isolated, and cloned, on which further analysis was conducted.73–75 More than 2000 mutations have been identified in the CFTR gene which the most notable of them are: p.Phe508del or c.1521_1523delCTT, G542X, and G551D.76–78 Each one of these mutations (based on the defect that occurs in the CFTR protein) leads to different forms of molecular pathways in the CF disease. Note that this difference may not be easily visible in clinical symptoms of patients. Therefore, in the same disease such as CF all patients with different mutations and, consequently, different types of the illness may have similar symptoms. In relation to the CF case, the G542X mutation leads to a relative reduction or a complete lack of protein synthesis in patients. On the other hand, in patients with p.Phe508del mutation (some sources have written this mutation as ΔF508del) although the protein is produced, it does not adequately reach the epithelial membrane surface. Finally, in patients with the G551D mutation, the protein is produced and placed in its proper position in the membrane, but it does not function normally.28,79,80 Therefore, each one of these mutations through different ways leads to the CF disease. Consequently, recognizing the correct and accurate type of mutation, and then categorizing the patients based on their specific mutations can lead to simple and better treatments of the disease. In 2003, a study was conducted by Wilschanski et al;79 they found that gentamicin can induce expression of CFTR gene and produce its protein; this interference led to symptomatic relief in patients carrying homozygous stop mutations (eg, G542X mutation). However, it has not been effective in patients who were homozygous for ΔF508 (p.Phe508del).79 Another example is the drug ivacaftor. This drug is designed to correct the function of the protein located in the cell membrane. Therefore, it is most effective in people who produced the CFTR protein and it reaches the cell surface (eg, patients with G551D mutation), but it is not effective or, in the best state, have a low effect in CF patients who generated CFTR protein but, it does not reach the cell surface (eg, those carrying p.Phe508del mutation).81,82 Accordingly, we can see how the understandings of genetic changes in patients can lead to designing precise therapies and medications for diseases according to the type of mutations.
Oncology
Cancer is a genetic disease which occurs due to changes in some genes involved in cell growth and division.83,84 Epigenetic disorder mechanisms are one of the causes of cancer. The most important of these changes is the DNA methylation, which leads to the spread of Helicobacter pylori and inflammatory processes followed by induction of DNA methylation disorder. Mutations and epigenetic changes are the two main agents of neoplasia.
Genetic changes that can lead to cancer can be inherited through parents or caused by environmental factors such as prolonged exposure to radiation or hazardous chemical materials. In both cases (inheritance and environment), irreversible alterations may occur in DNA, and these changes may lead to impairment in the function of some genes including proto-oncogenes, tumor suppressor genes, and DNA repair genes.85 As a response, the process of uncontrolled growth of the cell(s) can begin. As stated above, each person has a unique genetic content. This concept also applies to genetic variations. Indeed, each person has also a unique genetic variation. Herein, as we are focused on the changes that are related to cancer so, we can say that in cancer each individual has a unique genetic variation related to cancer. Hence, patients develop unique cancers with unique characteristics. Finally, we can conclude that designing specific treatment procedures for patients is necessary based on the specificity of cancer-related genetic alterations. In order to show the importance of identifying variations and its role in the application of precision medicine in cancer, some examples are reported as follows.
In ovarian cancer, patients with a mutation in the P53 gene (in particular, p53 overexpression as well as p53 missense mutations) exhibit resistance to some chemotherapy procedures such as chemotherapy via platinum.86–88 In other studies, it was observed that overexpression of collagen VI, collagen type XI alpha 1, and SUSD2 gene could play a role in resistance to cisplatin treatment.89–91 In relation to breast cancer, there are several examples of different therapeutic responses in patients. Mutations in heat shock proteins (Hsps) genes, and especially Hsp 70, can play a significant role in drug resistance.92 Overexpression of the HER2 (erbB-2), BARD1, and BRCA1 as well as genetic polymorphisms in CYP2D6 and ABCC2 can lead to drug resistance against tamoxifen in breast cancer patients.93–95 Some of these alterations have been identified. Undoubtedly, there are a number of variations that have yet to be discovered and their discovery will increase our insights on cancer. Therefore, through the identification of these alterations more effective treatments can be developed especially, for the subtypes of each cancer that show no response to current treatment. In previous studies, efforts have been made in this field.96 Consequently, the general prescription of chemotherapy drugs (especially for patients who are resistant to these drugs) will be prevented.
Altogether, the cases mentioned above constitute only a small part of the link between genetic variation and its effect on the resistance or susceptibility of cancer to existing drugs. There are many more examples, but the important concern is that in relation to cancer the mechanisms of resistance or sensitivity of the drug(s) have not been completely understood by now.97–100 Therefore, only based on similar clinical manifestations we cannot use a general treatment for people with the same situation such as cancer. Indeed, to achieve more satisfactory results, understanding the alterations and mechanisms affecting the treatment is essential, and would be promising for overcoming the obstacles existing in the cancer treatment process.
Conclusion
Today, some diseases are resistant to existing medications and treatments.101 Further, there has been no definitive treatment for some diseases by the time of writing this article.1,2,102–105 Therefore, it seems that we need to change the existing treatment procedures against some diseases, as sometimes insisting on attempts to treat a certain disease through existing methods not only cannot result in any improvement, but also may lead to side effects. For example, excessive use of certain chemotherapy drugs in people who are resistant to drugs not only does not result in effective treatment but it can also lead to serious side effects in patients. In order to design an accurate treatment and prevent the side effects of the drugs prescribed, the precision medicine is a field which can be very promising and as mentioned, it has been successful in some cases.
The study of the genome (genomics) and transcriptome (transcriptomics) is very important in discovering disease pathways and the design of precise and effective drugs. Increase insight into these areas can be helpful in treating diseases that are resistant to existing treatments. Development of nucleic acid sequencing methods and in particular the next-generation sequencing technologies is one of the most important tools for studying these levels. At the genome level, many studies have been conducted on the discovery of individual differences in a patient with the same condition. Most of the important studies are based on the human genome project, single nucleotide polymorphism detection,106 and genome wide association studies (GWAS).107 In relation to the transcriptome splicing, alternative splicing, and RNA sequencing are very important fields as many human genes are involved in the alternative splicing process and alterations in this stage are not easily detectable at the genome level.
In addition to the precision in drug prescription, another major difference between the precision medicine and conventional medicine is that conventional medicine often acts when a person is sick and does not have a preventive role while the precision medicine can also play a predictive and preventive role in the early stage of disease. For instance, before appearance of the symptoms of a disease in a person who seems to be predisposed, treatment can be initiated through finding related risk factors. In this case, healthy people who have cancer patients in their families can be informed about their health status through screening programs such as DNA or RNA (cDNA) sequencing, performed by next-generation sequencing (NGS) or other sequencing technologies. Examples include the role of BRCA genes mutations in preventing breast cancer. It is clear that achieve these goals, there will be problems that have been studied in detail in previous studies.108,109
If we want to summarize the precision medicine term in a short and comprehensive model, we might find the best description in a phrase from Dr. Mark A. Frye: “right drug, right dose, right patient”.110 Nevertheless, the modern precision medicine, as what we see in other sciences, is an interdisciplinary science, and it cannot attain its goals and achievements except utilizing all the existing potentials, especially through advances in genetic tools and techniques for identifying interrelationships between diseases and the genetic characteristics of patients as well as the ability to translating these data into clinical applications. Understanding the differences associated with a disease is possible at different levels, including the genome, transcriptome, proteome, and metabolome. In some cases, these alterations can only be detected at one of the levels mentioned, and not at all of them. Therefore, in order to achieve the desired goals related to information about all of the pathways and levels which affect the disease must be obtained. In the next step, we will need powerful tools and equipment to store and share a large amount of data. One of the great advances in this field is the use of mobile phones and apps technology to collect and share big data.111 Ultimately, we need the knowledge and expertise to translate the archived data into the clinical applications. However, we should not allow that the demand for these tools and equipment lead to forgetting one of the main goals of the healthcare systems (including precision medicine), ie, cheapness and affordability for low-income patients. Finally, it is hoped that one day all human beings will equally taste the fruits of the science tree whether patients who are living with enduring extreme hardships in inaccessible small villages in Asia and Africa or famous politicians and prosperous people who are living in the vicinity of advanced hospitals and reference laboratories.
Acknowledgment
The authors would like to thank to Dr. S.Azizi, Dr. Hosseinnia, and Mr. Zadrostam for their valuable support.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Wang Y, Deng W, Li N, et al. Combining immunotherapy and radiotherapy for cancer treatment: current challenges and future directions. Front Pharmacol. 2018;9:185. doi:10.3389/fphar.2018.00185
2. Khodadadian A, Hemmati-Dinarvand M, Kalantary-Charvadeh A, Ghobadi A, Mazaheri M. Candidate biomarkers for Parkinson’s disease. Biomed Pharmacother. 2018;104:699–704. doi:10.1016/j.biopha.2018.05.026
3. Miyasaki JM, Martin W, Suchowersky O, Weiner WJ, Lang AE. Practice parameter: initiation of treatment for Parkinson’s disease: an evidence-based review. Rep Qual Stand Subcommittee Am Acad Neurol. 2002;58(1):11–17.
4. Patrono C, Coller B, Dalen JE, FitzGerald GA. Platelet-active drugs: the relationships among dose, effectiveness, and side effects. Chest. 2001;119(1):S39. doi:10.1378/chest.119.1_suppl.39S
5. Hameed HMA, Islam MM, Chhotaray C, et al. Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-mycobacterium tuberculosis strains. Front Cell Infect Microbiol. 2018;8:114. doi:10.3389/fcimb.2018.00114
6. Wang L, Ma L, Xu F, et al. Role of long non-coding RNA in drug resistance in non-small cell lung cancer. Thorac Cancer. 2018.
7. Group CDPR. Gallbladder disease as a side effect of drugs influencing lipid metabolism experience in the coronary drug project. N Engl J Med. 1977;296(21):1185–1190. doi:10.1056/NEJM197705262962101
8. Sada K-E, Yamamura M, Harigai M, et al. Different responses to treatment across classified diseases and severities in Japanese patients with microscopic polyangiitis and granulomatosis with polyangiitis: a nationwide prospective inception cohort study. Arthritis Res Ther. 2015;17(1):305. doi:10.1186/s13075-015-0815-y
9. Rivenbark AG, O’Connor SM, Coleman WB. Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine. Am J Pathol. 2013;183(4):1113–1124. doi:10.1016/j.ajpath.2013.08.002
10. Gæde P, Vedel P, Larsen N, Jensen GV, Parving -H-H, Pedersen O. Multifactorial intervention and cardiovascular disease in patients with type 2 diabetes. N Engl J Med. 2003;348(5):383–393. doi:10.1056/NEJMoa021778
11. Kruskall MS, Leonard S, Klapholz H. Autologous blood donation during pregnancy: analysis of safety and blood use. Obstet Gynecol. 1987;70(6):938–941.
12. Gillespie TW, Hillyer CD. Blood donors and factors impacting the blood donation decision. Transfus Med Rev. 2002;16(2):115–130. doi:10.1053/tmrv.2002.31461
13. Marson FA, Bertuzzo CS, Ribeiro JD. Personalized or precision medicine? The example of cystic fibrosis. Front Pharmacol. 2017;8:390.
14. Bianco AM, Marcuzzi A, Zanin V, Girardelli M, Vuch J, Crovella S. Database tools in genetic diseases research. Genomics. 2013;101(2):75–85. doi:10.1016/j.ygeno.2012.11.001
15. Xu C, Wu K, Zhang JG, Shen H, Deng HW. Low‐, high‐coverage, and two‐stage DNA sequencing in the design of the genetic association study. Genet Epidemiol. 2017;41(3):187–197. doi:10.1002/gepi.22015
16. Xue Y, Lameijer E-W, Ye K, et al. Precision medicine: what challenges are we facing? Genomics Proteomics Bioinformatics. 2016;14(5):253–261. doi:10.1016/j.gpb.2016.10.001
17. Castellani C, Assael BM. Cystic fibrosis: a clinical view. Cell Mol Life Sci. 2017;74(1):129–140.
18. Marson FAL, Bertuzzo CS, Ribeiro JD. Personalized or precision medicine? The example of cystic fibrosis. Front Pharmacol. 2017;8:390.
19. Issa AM. Personalized medicine and the practice of medicine in the 21st century. McGill J Med. 2007;10(1):53.
20. Jameson JL, Longo DL. Precision medicine—personalized, problematic, and promising. Obstet Gynecol Surv. 2015;70(10):612–614. doi:10.1097/01.ogx.0000472121.21647.38
21. Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17(9):507. doi:10.1038/nrg.2016.86
22. Condren ME, Bradshaw MD. Ivacaftor: a novel gene-based therapeutic approach for cystic fibrosis. J Ped Pharmacol Ther. 2013;18(1):8–13. doi:10.5863/1551-6776-18.1.8
23. Davies JC, Wainwright CE, Canny GJ, et al. Efficacy and safety of ivacaftor in patients aged 6 to 11 years with cystic fibrosis with a G551D mutation. Am J Respir Crit Care Med. 2013;187(11):1219–1225. doi:10.1164/rccm.201301-0153OC
24. Rowe SM, Heltshe SL, Gonska T, et al. Clinical mechanism of the cystic fibrosis transmembrane conductance regulator potentiator ivacaftor in G551D-mediated cystic fibrosis. Am J Respir Crit Care Med. 2014;190(2):175–184. doi:10.1164/rccm.201404-0703OC
25. Wainwright CE, Elborn JS, Ramsey BW, et al. Lumacaftor–ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med. 2015;373(3):220–231. doi:10.1056/NEJMoa1409547
26. Boyle MP, Bell SC, Konstan MW, et al. A CFTR corrector (lumacaftor) and a CFTR potentiator (ivacaftor) for treatment of patients with cystic fibrosis who have a phe508del CFTR mutation: a Phase 2 randomised controlled trial. Lancet Respir Med. 2014;2(7):527–538. doi:10.1016/S2213-2600(14)70132-8
27. Duster T. Race and Reification in Science. American Association for the Advancement of Science; 2005.
28. Turnpenny PD, Ellard S. Emery’s Elements of Medical Genetics. Vol. 15. Elsevier; 2017.
29. Litman T. Personalized medicine—concepts, technologies, and applications in inflammatory skin diseases. Apmis. 2019;127(5):386–424.
30. Kranzler HR, Smith RV, Schnoll R, Moustafa A, Greenstreet-Akman E. Precision medicine and pharmacogenetics: what does oncology have that addiction medicine does not? Addiction. 2017;112(12):2086–2094.
31. Morley JE, Anker SD. Myopenia and precision (P4) medicine. J Cachexia Sarcopenia Muscle. 2017;8(6):857–863.
32. Personalized/Precision Medicine. Wiley StatsRef: Statistics Reference Online. p. 1–7.
33. Morgan S, Grootendorst P, Lexchin J, Cunningham C, Greyson D. The cost of drug development: a systematic review. Health Policy (New York). 2011;100(1):4–17. doi:10.1016/j.healthpol.2010.12.002
34. Miller JE, Korn D, Ross JS. Clinical trial registration, reporting, publication and FDAAA compliance: a cross-sectional analysis and ranking of new drugs approved by the FDA in 2012. BMJ Open. 2015;5(11):e009758. doi:10.1136/bmjopen-2015-009758
35. Wadman M. Experts call for active surveillance of drug safety. Nature. 2007;446:358–359. doi:10.1038/446358b
36. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi:10.1056/NEJMp1500523
37. Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17(3):297. doi:10.1038/nm.2323
38. Eckhard U, Marino G, Butler GS, Overall CM. Positional proteomics in the era of the human proteome project on the doorstep of precision medicine. Biochimie. 2016;122:110–118. doi:10.1016/j.biochi.2015.10.018
39. Duarte TT, Spencer CT. Personalized proteomics: the future of precision medicine. Proteomes. 2016;4(4):29. doi:10.3390/proteomes4040029
40. NMR-based pharmacometabonomics: a new approach to personalized medicine. eMagRes. 197–208.
41. Watson JD, Crick FHC. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737. doi:10.1038/171737a0
42. Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94(3):441–448. doi:10.1016/0022-2836(75)90213-2
43. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74(12):5463–5467. doi:10.1073/pnas.74.12.5463
44. Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci. 1977;74(2):560–564. doi:10.1073/pnas.74.2.560
45. Saiki RK, Gelfand DH, Stoffel S, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239(4839):487–491. doi:10.1126/science.239.4839.487
46. Eisen JA. Badomics words and the power and peril of the ome-meme. GigaScience. 2012;1(1):1–3. doi:10.1186/2047-217X-1-6
47. Zelenin A, Rodionov A, Bolsheva N, Badaeva E, Genome: MO. Origins and evolution of the term. Mol Biol. 2016;50(4):542–550. doi:10.1134/S0026893316040178
48. Lynch M, Walsh B. The Origins of Genome Architecture. Sunderland (MA): Sinauer Associates; 2007.
49. Brosius J. The fragmented gene. Ann N Y Acad Sci. 2009;1178(1):186–193. doi:10.1111/j.1749-6632.2009.05004.x
50. Griffiths AJF GW, Miller JH, et al. Modern Genetic Analysis. New York: W. H. Freeman; 1999. The Nature of Genomes. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21342/.
51. Robinson R Genetics: Macmillan Reference Lib; 2002.
52. Klug WS, Cummings MR, Palladino MA, Spencer CA. Concepts of Genetics. Pearson Education; 2012.
53. Roth SC. What is genomic medicine? J Med Libr Assoc. 2019;107(3):442–448. doi:10.5195/jmla.2019.604
54. Collins FS. Medical and societal consequences of the human genome project. N Engl J Med. 1999;341(1):28–37. doi:10.1056/NEJM199907013410106
55. Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the US human genome project: 1998-2003. science. 1998;282(5389):682–689. doi:10.1126/science.282.5389.682
56. Consortium IH. A haplotype map of the human genome. Nature. 2005;437(7063):1299.
57. Ball MP, Thakuria JV, Zaranek AW, et al. A public resource facilitating clinical use of genomes. Proc Natl Acad Sci. 2012;109(30):11920–11927. doi:10.1073/pnas.1201904109
58. Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366(6):489–491. doi:10.1056/NEJMp1114866
59. Lowe R, Shirley N, Bleackley M, Dolan S, Shafee TJP. Transcriptomics technologies. 2017;13(5):e1005457.
60. Clark K, P K, Tatusova T, et al. BioProject. 2013 Apr 28 [Updated 2013 Nov 11]. In: The NCBI Handbook [Internet].
61. Lowe R, Shirley N, Bleackley M, Dolan S, Shafee TJP. Transcriptomics technologies. PLoS Comp Biol. 2017;13:5.
62. Sharp PA. Split genes and RNA splicing. Cell. 1994;77(6):805–815. doi:10.1016/0092-8674(94)90130-9
63. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413. doi:10.1038/ng.259
64. Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2015;17:19. doi:10.1038/nrg.2015.3
65. Mattick JS, Makunin IVJ. Non-coding RNA. Human Mol Genetics. 2006;15(suppl_1):R17–R29.
66. Moein S, Vaghari-Tabari M, Qujeq D, Majidinia M, Nabavi SM, Yousefi B. MiRNAs and inflammatory bowel disease: an interesting new story. J Cell Physiol. 2019;234(4):3277–3293.
67. Anvarnia A, Mohaddes-Gharamaleki F, Asadi M, Akbari M, Yousefi B, Shanehbandi D. Dysregulated microRNAs in colorectal carcinogenesis: new insight to cell survival and apoptosis regulation. Journal of Cellular Physiology. 2019;234(12):21683–21693. doi:10.1002/jcp.28872
68. Consortium IWP. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764.
69. Pirmohamed M. Pharmacogenetics and pharmacogenomics. Br J Clin Pharmacol. 2001;52(4):345–347. doi:10.1046/j.0306-5251.2001.01498.x
70. Zaiou M, El Amri H. Cardiovascular pharmacogenetics: a promise for genomically-guided therapy and personalized medicine. Clin Genetics. 2017;91(3):355–370.
71. Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. science. 1999;286(5439):487–491. doi:10.1126/science.286.5439.487
72. Estivill X, Farrall M, Scambler PJ, et al. A candidate for the cystic fibrosis locus isolated by selection for methylation-free islands. Nature. 1987;326(6116):840. doi:10.1038/326840a0
73. Rommens JM, Iannuzzi MC, Kerem B-S, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245(4922):1059–1065. doi:10.1126/science.2772657
74. Riordan JR, Rommens JM, Kerem B-S, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245(4922):1066–1073. doi:10.1126/science.2475911
75. Kerem B-S, Rommens JM, Buchanan JA, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245(4922):1073–1080. doi:10.1126/science.2570460
76. Berwouts S, Morris MA, Girodon E, Schwarz M, Stuhrmann M, Dequeker E. Mutation nomenclature in practice: findings and recommendations from the cystic fibrosis external quality assessment scheme. Hum Mutat. 2011;32(11):1197–1203. doi:10.1002/humu.21569
77. Cordovado S, Hendrix M, Greene C, et al. CFTR mutation analysis and haplotype associations in CF patients. Mol Genet Metab. 2012;105(2):249–254. doi:10.1016/j.ymgme.2011.10.013
78. Trouvé P, Kerbiriou M, Teng L, et al. G551D-CFTR needs more bound actin than wild-type CFTR to maintain its presence in plasma membranes. Cell Biol Int. 2015;39(8):978–985.
79. Wilschanski M, Yahav Y, Yaacov Y, et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med. 2003;349(15):1433–1441. doi:10.1056/NEJMoa022170
80. Kristidis P, Bozon D, Corey M, et al. Genetic determination of exocrine pancreatic function in cystic fibrosis. Am J Hum Genet. 1992;50(6):1178.
81. Ramsey BW, Davies J, McElvaney NG, et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med. 2011;365(18):1663–1672. doi:10.1056/NEJMoa1105185
82. Brodlie M, Haq IJ, Roberts K, Elborn JS. Targeted therapies to improve CFTR function in cystic fibrosis. Genome Med. 2015;7(1):101. doi:10.1186/s13073-015-0223-6
83. Yousefi B, Mohammadlou M, Abdollahi M, et al. Epigenetic changes in gastric cancer induction by Helicobacter pylori. Journal of Cellular Physiology. 2019;234(12):21770–21784. doi:10.1002/jcp.28925
84. Jahanban-Esfahlan R, de la Guardia M, Ahmadi D, Yousefi B. Modulating tumor hypoxia by nanomedicine for effective cancer therapy. J Cell Physiol. 2018;233(3):2019–2031.
85. Valizadeh A, Majidinia M, Samadi-Kafil H, Yousefi M, Yousefi B. The roles of signaling pathways in liver repair and regeneration. J Cell Physiol. 2019;234(9):14966–14974.
86. Reles A, Wen WH, Schmider A, et al. Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer. Clin Cancer Res. 2001;7(10):2984–2997.
87. Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3(7):502. doi:10.1038/nrc1123
88. Righetti SC, Della Torre G, Pilotti S, et al. A comparative study of p53 gene mutations, protein accumulation, and response to cisplatin-based chemotherapy in advanced ovarian carcinoma. Cancer Res. 1996;56(4):689–693.
89. Xu Y, Miao C, Jin C, et al. SUSD2 promotes cancer metastasis and confers cisplatin resistance in high grade serous ovarian cancer. Exp Cell Res. 2018;363(2):160–170. doi:10.1016/j.yexcr.2017.12.029
90. Sherman-Baust CA, Weeraratna AT, Rangel LB, et al. Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell. 2003;3(4):377–386. doi:10.1016/S1535-6108(03)00058-8
91. Rada M, Nallanthighal S, Cha J, et al. Inhibitor of apoptosis proteins (IAPs) mediate collagen type XI alpha 1-driven cisplatin resistance in ovarian cancer. Oncogene. 2018;37(35):4809–4820. doi:10.1038/s41388-018-0297-x
92. Vargas‐Roig LM, Gago FE, Tello O, Aznar JC, Ciocca DR. Heat shock protein expression and drug resistance in breast cancer patients treated with induction chemotherapy. Int J Cancer. 1998;79(5):468–475. doi:10.1002/(SICI)1097-0215(19981023)79:5<468::AID-IJC4>3.0.CO;2-Z
93. Kurokawa H, Lenferink AE, Simpson JF, et al. Inhibition of HER2/neu (erbB-2) and mitogen-activated protein kinases enhances tamoxifen action against HER2-overexpressing, tamoxifen-resistant breast cancer cells. Cancer Res. 2000;60(20):5887–5894.
94. Zhu Y, Liu Y, Zhang C, et al. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat Commun. 2018;9(1):1595. doi:10.1038/s41467-018-03951-0
95. Kiyotani K, Mushiroda T, Imamura CK, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28(8):1287. doi:10.1200/JCO.2009.25.7246
96. Áyen Á, Jiménez Martínez Y, Marchal JA, Boulaiz HJI. Recent progress in gene therapy for ovarian cancer. Int J Mol Sci. 2018;19(7):1930.
97. Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53(1):615–627. doi:10.1146/annurev.med.53.082901.103929
98. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714. doi:10.1038/nrc3599
99. Norouzi‐Barough L, Sarookhani MR, Sharifi M, Moghbelinejad S, Jangjoo S, Salehi R. Molecular mechanisms of drug resistance in ovarian cancer. J Cell Physiol. 2018;233(6):4546–4562.
100. Mihanfar A, Aghazadeh Attari J, Mohebbi I, et al. Ovarian cancer stem cell: a potential therapeutic target for overcoming multidrug resistance. 2019;234(4):3238–3253.
101. Hasanifard L, Sheervalilou R, Majidinia M, Yousefi B. New insights into the roles and regulation of SphK2 as a therapeutic target in cancer chemoresistance. J Cell Physiol. 2019;234(6):8162–8181.
102. Budd Haeberlein SL, Harris TJ. Promising targets for the treatment of neurodegenerative diseases. Clin Pharmacol Ther. 2015;98(5):492–501. doi:10.1002/cpt.195
103. Loh KP, Lin PJ, Uth J, Quist M, Klepin H, Mustian K. Exercise for managing cancer- and treatment-related side effects in older adults. J Immunol Res. 2018.
104. Lukasiewicz K, Fol M. Microorganisms in the treatment of cancer: advantages and limitations. J Immunol Res. 2018;2018:2397808.
105. Smith DK, He M, Zhang CL, Zheng JC. The therapeutic potential of cell identity reprogramming for the treatment of aging-related neurodegenerative disorders. Prog Neurobiol. 2017;157:212–229. doi:10.1016/j.pneurobio.2016.01.006
106. Jain K. Personalized medicine. Curr Opin Mol Ther. 2002;4(6):548–558.
107. McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356. doi:10.1038/nrg2344
108. Manzoni C, Kia DA, Vandrovcova J, et al. Genome transcriptome proteome. 2018;19(2):286–302.
109. Kotsopoulos JJC. BRCA mutations and breast cancer prevention. Cancers. 2018;10(12):524.
110. clinic M. Right drug, right dose, right patient 2014. Available from: https://www.mayoclinic.org/medical-professionals/clinical-updates/psychiatry-psychology/right-drug-right-dose-right-patient.
111. Council NR. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. National Academies Press; 2011.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.