Back to Journals » Infection and Drug Resistance » Volume 15
Genomic Characterization of ESBL/AmpC-Producing Escherichia coli in Stray Dogs Sheltered in Yangzhou, China
Authors Sun L, Meng N, Wang Z, Hong J , Dai Y, Wang Z, Wang J, Jiao X
Received 18 November 2022
Accepted for publication 20 December 2022
Published 28 December 2022 Volume 2022:15 Pages 7741—7750
DOI https://doi.org/10.2147/IDR.S397872
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Lin Sun,1,2,* Nan Meng,1,* Zecheng Wang,1 Jiaxin Hong,1 Yuqi Dai,1 Zhenyu Wang,1 Jing Wang,1,2 Xinan Jiao1,2
1Jiangsu Key Laboratory of Zoonosis/Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, People’s Republic of China; 2Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jing Wang; Xinan Jiao, Email [email protected]; [email protected]
Purpose: Limited data are available on the prevalence and antimicrobial resistance of extended spectrum β-lactamase- (ESBL) and AmpC β-lactamase-producing Escherichia coli in stray dogs. We aimed to investigate the genomic characteristics of ESBL/AmpC-producing E. coli isolated from stray dogs sheltered in Yangzhou, China.
Methods: We collected 156 samples including 115 fecal swabs, 35 kennel floor swabs, two breeder hand and shoe sole swabs, and four feed samples. The isolates were tested for resistance by antimicrobial susceptibility testing and further analyzed for cefotaxime-resistant E. coli isolates by whole genome sequencing.
Results: We identified 80 cefotaxime-resistant E. coli isolates (51.3%), 59 isolates (73.8%) from feces and 21 (26.2%) from the environment. Whole-genome sequencing analysis showed that blaCTX-M-15 (n=30) and blaCTX-M-55 (n=29) were the most prevalent genotypes. Two isolates only carried the AmpC β-lactamase gene blaCMY-2; one isolate had a combination of AmpC β-lactamase gene blaDHA-1 and ESBL β-lactamase gene blaCTX-M-14. Other important resistance genes such as blaOXA-10, blaTEM-1B, blaTEM-135, blaTEM-106, tet(A), qnrS1, qnrB4, and oqxAB were also detected. The serotype combination was highly abundant, with O10:H25 predominating (n=12). Most cefotaxime-resistant E. coli isolates belonged to phylogroup A (62.5%, n=50), followed by phylogroup B1 (26.3%, n=21). Thirty different sequence types (STs) and 27 distinct plasmid replicons were identified, among which ST2325 (n=12) and IncFII (n=38) was the most frequent ST and plasmid, respectively. ESBL/AmpC-producing isolates were divided into four major clades; clade IV was the primary lineage containing 37 isolates from feces and 13 from the environment. Three high-risk E. coli clone ST23 strains and one ST10 strain belonged to clades III and IV, respectively.
Conclusion: Our study provides a comprehensive overview of resistance profiles and genomic characteristics in ESBL/AmpC-producing E. coli and highlights the possible role of stray dogs as an antibiotic resistance gene reservoir.
Keywords: Escherichia coli, ESBL/AmpC, stray dogs, antimicrobial resistance, whole genome sequencing
Plain Language Summary
- High prevalence (51.3%) of ESBL/AmpC-producing E. coli was found among stray dogs and in their surrounding environment.
- blaCTX-M-15 and blaCTX-M-55 were the most prevalent genotypes.
- Stray dogs could be an antibiotic resistance gene reservoir, representing a significant risk to public health.
Introduction
Antimicrobial resistance (AMR) has been highlighted as a serious threat to public health worldwide. Owing to their high efficacy and low toxicity, cephalosporins are widely used in human clinical therapy and veterinary medicine.1 However, the intensive use of cephalosporins has led to the widespread dissemination of cephalosporins resistance in bacteria. An important mechanism of resistance to cephalosporins in Escherichia coli is the production of extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases (AmpCs).2
ESBLs mainly contain sulfhydryl variable (SHV), temoniera (TEM), oxacillinase (OXA), and cefotaximases (CTX-M). Currently, CTX-M type ESBL has become the most common ESBL worldwide, and the most common host is E. coli.3 Among the different CTX-M subtypes, CTX-M-1 and CTX-M-9 groups are predominant worldwide.4,5 High levels of AmpC β-lactamases via overexpression of the chromosomally encoded AmpC gene (c-AmpC) and/or presence of plasmid-mediated ampC gene (p-AmpC) are also important mechanisms mediating resistance to cephalosporins.6
ESBL/AmpC-encoding E. coli isolates have been widely reported worldwide, and with the assistance of mobile elements, have become one of the fastest-transmitting resistant pathogens in humans, animals, and the environment.7
Human-animal transmission of ESBL/AmpC-producing bacteria was previously described, non-human sources are important hosts involved in further spread of bacterial infections. Unlike wild animals or food animals, which have received widespread attention, the role of animals with a sheltered or stray background as important carriers of drug-resistant bacteria has been underestimated.8–10 The close contact of these animals with humans in some cases provides a suitable bridge for cross-species transmission of multidrug-resistant (MDR) bacteria. In fact, AMR surveillance of stray animals (mainly dogs) is critical because they may play a role in the transmission of MDR bacteria to humans via direct or indirect contact when sharing the same environment.11 Potential risk factors for ESBL/AmpC carriage in stray dogs are free movement across large areas, scavenging of food remains and human/animal waste, lack of sanitation, and exposure to the environmental resistome.12
Currently, studies on ESBL/AmpC-carrying E. coli in stray dogs are scarce, and the potential transmission of ESBL/AmpC-carrying E. coli from stray dogs to the surrounding environment or humans has not been investigated in detail. Therefore, we aimed to investigate the prevalence and characterization of ESBL/AmpC-producing E. coli isolates from stray dogs and related environments and to elucidate their genetic diversity and dissemination mechanisms.
Materials and Methods
Sample Collection and Bacterial Isolation
A total of 156 samples were collected from Yangzhou Canine Inspection and Quarantine Institute in July 2021, including 115 fecal swabs, 35 kennel floor swabs, two breeder hand and shoe sole swabs, and four feed samples. The samples were pre-enriched in 2 mL of Luria-Bertani broth at 37 °C, 180 rpm for 24 hours. Ten-microliter aliquots of each enriched samples were inoculated on MacConkey agar plates supplemented with 2 mg/L cefotaxime and incubated at 37 °C overnight. One E. coli isolate per plate was selected for further purification and identified using Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed using agar dilution or broth microdilution methods (limited to colistin and tigecycline). The tested antibiotics included ampicillin, cefazolin, cefotaxime, meropenem, gentamicin, amikacin, streptomycin, tetracycline, tigecycline, nalidixic acid, ciprofloxacin, colistin, trimethoprim-sulfamethoxazole, chloramphenicol, florfenicol, and fosfomycin. Results were interpreted according to the 2020 Clinical and Laboratory Standards Institute guidelines, except the breakpoints of streptomycin, tigecycline, and florfenicol were interpreted using European Committee on Antimicrobial Susceptibility Testing guidelines (https://www.eucast.org/). E. coli ATCC 25922 was used as the quality control strain.
Whole-Genome Sequencing and Analysis
All cefotaxime-resistant E. coli isolates were sequenced using the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA), and at least 100-fold coverage of raw reads were collected. The raw sequence data were assembled into contigs using SPAdes 3.10.0.13 Species identification was also performed based on the genomic data (https://pathogen.watch/). Sequence types (STs) were identified using the E. coli Multilocus Sequence Typing (MLST) database (https://bigsdb.pasteur.fr/ecoli/). Antimicrobial resistance genes and plasmid replicons were analyzed via the Center for Genomic Epidemiology pipeline (https://cge.cbs.dtu.dk/). In silico E. coli phylogroup stratification was performed using ClermonTyping (http://clermontyping.iame-research.center/).14 The phylogenetic tree based on core genome single nucleotide polymorphisms (SNPs) was generated using ParSNP v. 1.2 (https://github.com/marbl/parsnp).15 ChiPlot (http://chiplot.online/#) was used to visualize the phylogenetic tree.
Results
Prevalence of Cefotaxime-Resistant E. coli Strains and Antimicrobial Susceptibility
Of total 156 samples, 80 cefotaxime-resistant E. coli isolates (51.3%) were identified, including 59 isolates (73.8%) from feces and 21 isolates (26.2%) from the environment. The environmental isolates included 20 isolates from floor swabs and one from breeder shoe sole swabs.
Eighty cefotaxime-resistant E. coli isolates showed differential resistance to 16 antibiotics (Table 1). All isolates were resistant to ampicillin and cefotaxime. High resistance rates (>60%) were observed for tetracycline and trimethoprim-sulfamethoxazole. However, all the isolates were susceptible to meropenem, amikacin, colistin, and tigecycline. Most cefotaxime-resistant E. coli isolates, including those of feces origin (57/59, 96.6%) and environmental origin (20/21, 95.2%), exhibited MDR phenotypes (≥3 antimicrobial classes) (Figure 1). The isolates from feces had more complex drug resistance phenotypes than those from the environment, and two isolates from feces showed resistance to 12 agents, with a resistance spectrum of ampicillin, cefazolin, cefotaxime, gentamicin, streptomycin, tetracycline, nalidixic acid, ciprofloxacin, trimethoprim-sulfamethoxazole, chloramphenicol, florfenicol, and fosfomycin.
 |
Table 1 Antimicrobial Resistance Profiles of 80 Cefotaxime-Resistant Escherichia coli Isolates |
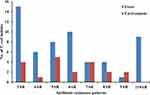 |
Figure 1 Antimicrobial resistance (AR) patterns of cefotaxime-resistant E. coli isolates from stray dog feces and the environment. |
Distribution of Antimicrobial Resistance Genes in Cefotaxime-Resistant E. coli
In total, 43 different resistance determinants were identified from 80 cefotaxime-resistant E. coli isolates, which was consistent with their MDR phenotypes. Eight blaCTX-M variants were identified among 78 isolates (97.5%), namely, blaCTX-M-15 (n=30), blaCTX-M-55 (n=29), blaCTX-M-14 (n=9), blaCTX-M-132 (n=4), blaCTX-M-27 (n=2), blaCTX-M-65 (n=2), blaCTX-M-3 (n=1), and blaCTX-M-102 (n=1) (Figure 2). Additionally, two isolates only carried the AmpC β-lactamase gene blaCMY-2. One isolate had a combination of AmpC β-lactamase gene blaDHA-1 and ESBL β-lactamase gene blaCTX-M-14. Other β-lactamase genes were also detected, such as blaOXA-10 (n=2) and TEM-type genes (30.0%, 24/80), including blaTEM-1B (n=16), blaTEM-135 (n=4), and blaTEM-106 (n=3).
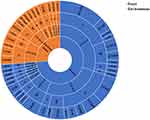 |
Figure 2 Distribution of ESBL/AmpC β-lactamase genes, sequence types (STs), and phylogroups in E. coli isolates. |
In addition to β-lactamase, genes accounting for resistance to aminoglycosides, trimethoprim, sulfonamides, tetracyclines, macrolides, phenicols, rifampicin, Fosfomycin, and fluoroquinolones were also identified (Figure 3). The tet(A) gene, which confers resistance to tetracycline, was detected in 57 isolates (71.3%). Plasmid-mediated quinolone resistance gene qnrS1 was detected in 57 isolates (71.3%); qnrB4 and oqxAB were also found in one isolate (1.2%) and two isolates (2.5%), respectively. The phenicol resistance gene floR (32.5%, 26/80), the sulfonamide resistance gene sul2 (37.5%, 30/80), the aminoglycoside resistance gene strAB (35.0%, 28/80), the trimethoprim resistance gene drfA14 (21.3%, 17/80), and the macrolide resistance gene mph(A) (22.5%, 18/80) were also identified.
Phylogroup, Multilocus Sequence Typing, and Serotyping of Cefotaxime-Resistant E. coli
Most cefotaxime-resistant E. coli isolates belonged to phylogroup A (62.5%, n=50), followed by phylogroup B1 (26.3%, n=21) (Figure 2). The remaining strains belonged to phylogroup C (3.7%, n=3), D (3.7%, n=3), B2 (2.5%, n=2), and F (1.3%, n=1). Most of the isolates from stray dog feces (46.3%, n=37) and the environment (16.3%, n=13) belonged to phylogroup A.
In silico multilocus sequence typing revealed 30 different STs among 80 cefotaxime-resistant E. coli isolates (Figure 2). Among them, the most frequent was ST2325 (n=12), followed by ST165 (n=8), ST48 (n=7), ST224 (n=5), ST100 (n=4), ST641 (n=4), ST2473 (n=4), ST6050 (n=4), ST13655 (n=4), ST23 (n=3), ST13659 (n=3), ST38 (n=2), ST746 (n=2), and ST1638 (n=2). The remaining isolates belonged to a variety of ST types (Figure 2). Additionally, two isolates belonged to phylogenetic group B2, isolated from stray dog feces, with MLST types ST73 and ST961, respectively. Three isolates of phylogenetic group D were derived from the environment (ST38) and feces (ST38 and ST349), respectively (Figure 2). The blaCTX-M-15-positive isolates were distributed across 10 different ST types, including ST38 and ST349. Twelve ST types were shared between fecal and environmental isolates and mainly encoded CTX-M-15 and CTX-M-55.
In serotype analysis, a total of 29 different serotypes were identified in 80 isolates. The serotype combination was highly abundant, with O10:H25 being predominant (n=12). O-serotypes are more abundant than flagellar serotypes (Figure 3).
Plasmid Replicons in Cefotaxime-Resistant E. coli
Plasmid replicon analysis revealed that 27 distinct plasmid replicons were identified in 80 cefotaxime-resistant E. coli isolates. As shown in Figure 3, IncF was the most common replicon type, including IncFII (n=38), IncFIA (n=8), and IncFIB (n=34). Other replicons such as IncI, IncHI, IncN, IncX, IncY, and Col-like plasmid (for example, Col440I, ColpVC) were also detected. In total, 56.3% isolates (n=45) carried two or more different plasmid replicons, and 17.5% (n = 14) did not harbor any detectable plasmid replicons.
Phylogenetic Analysis of Cefotaxime-Resistant E. coli Strains
To explore the relationship among the 80 cefotaxime-resistant E. coli isolates, we constructed a phylogenetic tree based on core genome SNPs. All ESBL-producing isolates were divided into four major clades, namely I (n=3, 3.75%), II (n=3, 3.75%), III (n=24, 30.00%), and IV (n=50, 62.50%) (Figure 3). Clade I contained three E. coli isolates from feces; the other clades contained both fecal and environmental isolates. Clade IV was the primary lineage, containing 37 isolates from feces and 13 isolates from the environment. ST2325, ST48, and ST165 were the predominant STs in this clade; one high-risk clone ST10 strain was designated as belonging to clade IV. Clade III was the second most prevalent lineage; ST224, ST641, and ST2473 were the predominant STs in this clade. Three high-risk clone ST23 strains belonged to clade III.
Discussion
Although the seriousness of antibiotic resistance has been recognized and certain interventions have been implemented to prevent the spread of antibiotic resistance, the problem is still growing. The relevance and role of AMR dissemination transcends the human context and is closely related to the human–animal–environment interface.16,17 As a bacterial host species that is closely related to the human living environment, stray dogs serve as a suitable bridge for the interspecies transmission of antimicrobial-resistant bacteria owing to their close contact with humans.
In the present study, high carriage (51.3%) of cefotaxime-resistant E. coli in stray dogs and shelters was reported. Apart from resistance to cefotaxime, the isolates showed resistance to ampicillin (100%), followed by high resistance to tetracycline and trimethoprim-sulfamethoxazole. This is consistent with the findings of many previous studies on companion animals and livestock18,19 and may be because β-lactams and tetracyclines are commonly used antibiotics in human and veterinary settings. The findings also suggest that resistance is broadly distributed across a wide range of environmental scenarios. Additionally, various rates of resistance to antibiotics such as quinolones (nalidixic acid and ciprofloxacin), gentamicin, and streptomycin, were observed in our isolated strains. These drugs are first-line antibiotics used to treat common infections and are critical to humans. The current treatment of ESBL Enterobacteriaceae infections, based on their susceptibility to drugs, is still preferred to carbapenems, and when carbapenems are not effective, colistin and tigecycline may be considered, but the treatment of the infection itself is complex and requires careful evaluation.20
Nearly all isolates (96.3%) exhibited multidrug-resistant phenotypes, with 11.3% of isolates showing resistance to more than 10 antibiotics. A previous study reported higher rates of drug resistance and multidrug resistance in E. coli strains from kennel dogs than those observed in strains from individually bred dogs.21 It is hypothesized that stray dogs themselves may carry multidrug-resistant bacteria. During shelter periods, dogs are exposed to poor sanitation and group housing, as well as other dogs and their bacteria-carrying feces, which are likely to contain multiple resistance genes that can be transmitted horizontally. Closely related extended-spectrum cephalosporin-resistant E. coli isolates have been found in dogs and humans sharing a shelter environment, suggesting clonal interspecies or dog-to-dog cross-transmission.22
In this study, the most important ESBL enzyme was the CTX-M enzyme, and eight different CTX-M types were detected, indicating a high diversity of blaCTX-M in E. coli isolates from stray dogs. These CTX-M variants were mainly clustered in the CTX-M-1 and CTX-M-9 groups, consistent with previous reports on the global prevalence of CTX-M genes.3,23 The most commonly detected genes in this study were blaCTX-M-15 and blaCTX-M-55, which are currently the most common types detected in pets and other animals in China and many other countries.24,25 Several variants (eg, CTX-M-65, CTX-M-14, and CTX-M-3) are also very common in E. coli isolated from humans.26,27 Similarly, CMY-2, the most prevalent pAmpC β-lactamase identified in this study, has been previously reported in E. coli from companion animals and stray dogs.8,28 Additionally, blaDHA-1 and blaCTX-M-14 genes were identified in the same strain. DHA-1 pAmpC β-lactamase was initially very common in human isolates; this gradually became more prevalent in companion animals in Europe, Korea, and other locations.29 Our study showed that the ESBL/AmpC-encoding genes in E. coli strains isolated from stray dogs (shelter/dog breeder) are becoming more diverse, posing a potential threat to humans.
Unlike previous reports, we did not find E. coli ST131 and ST410 clones with a high risk of international transmission.30,31 Results of molecular typing indicated that CTX-M-producing isolates were clonally diverse; other important E. coli genotypes identified in our study were ST10 group and ST38, isolated from both the environment and feces. Among them, the ST10 group (including ST10, ST48, and ST744) has been reported to be associated with urinary tract infections and sepsis in poultry and humans.32 Additionally, although most isolates were distributed in phylogenetic groups A and B1, a small number of isolates from phylogenetic group B2 and D were also identified. This is usually called virulence phylogeny, and most of these isolates contained more virulence factors.33 ST38 (belonging to phylogenetic group D) is usually considered an enteroaggregative E. coli strain and is frequently associated with urinary tract infections.34 In this study, two ST38 E. coli strains were isolated from fecal and environmental samples and showed a close phylogenetic relationship with the same resistance genes and IncFIB plasmids. Additionally, other ST types (such as ST48 and ST100) from different sources exist, but these contain the same resistance genes and plasmid replicons, indicating clonal transmission between different sources in the shelter. Simultaneously, highly diverse clones suggest the importance of horizontal transfer in the spread of resistance genes. Most blaCTX-M genes are associated with some well-characterized plasmids. For example, blaCTX-M-15 in the internationally prevalent clone ST131, is usually associated with IncF plasmids, which are vectors for blaCTX-M-14 dissemination among E. coli in Spain.35–37
The global prevalence of ESBL-E. coli is estimated to be relatively low in dogs compared to livestock or humans, but studies nonetheless reflect the existence of irrational antibiotic use.38 Shelter animals are likely to be an unusual source of infection for shelter staff or potential adopters or even families, and when infected with disease can cause a variety of problems for humans, such as length of hospitalization, mortality, and medical costs.39 Now that global data on the prevalence of ESBL-E. coli in stray dogs and cats are under reported, we still hope this study will serve as a wake-up call to improve public health.
We have to admit that there are some limitations in our study. The samples were small-scale, and only collected from Yangzhou City. Therefore more large-scale studies from different regions are needed.
Conclusion
In conclusion, our study demonstrated a high prevalence of ESBL- and pAmpC-producing E. coli among stray dogs in China. Owing to the complexity of the stray dog background, resulting in genetic diversity, further research is required to determine the spread of AMR genes in this population and their association with humans. It is critical to strengthen long-term monitoring, control the use of antibiotics, and reduce the risk of transmission of MDR bacteria among stray dogs.
Data Sharing Statement
The genome sequences in this study have been deposited into NCBI GenBank under BioProject PRJNA880799.
Ethical Statement
All the experimental procedures were approved by the Committee on the Ethics of Animal Experiments of Yangzhou University, Yangzhou, China, in accordance with Chinese legislations on animal experimentation.
Acknowledgment
We thank Analisa Avila, MPH, ELS, of Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the language of a draft of this manuscript.
Funding
This work was supported by the fifth phase of the “333 Project” scientific research project in Jiangsu Province (BRA2020002) and the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD) and Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX21_1597).
Disclosure
The authors report no conflicts of interest in this work.
References
1. European Food Safety A, European Centre for Disease P, Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020;18(3):e06007. doi:10.2903/j.efsa.2020.6007
2. Moland ES, Hanson ND, Black JA, Hossain A, Song W, Thomson KS. Prevalence of newer beta-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J Clin Microbiol. 2006;44(9):3318–3324. doi:10.1128/JCM.00756-06
3. Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–2155. doi:10.1093/jac/dkx146
4. Giufre M, Mazzolini E, Cerquetti M, Brusaferro S; Group CCMO-HE-pEcS. Extended-spectrum beta-lactamase-producing Escherichia coli from extraintestinal infections in humans and from food-producing animals in Italy: a ‘One Health’ study. Int J Antimicrob Agents. 2021;58(5):106433. doi:10.1016/j.ijantimicag.2021.106433
5. Kaesbohrer A, Bakran-Lebl K, Irrgang A, et al. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet Microbiol. 2019;233:52–60. doi:10.1016/j.vetmic.2019.03.025
6. Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect. 2012;18(7):646–655. doi:10.1111/j.1469-0691.2012.03850.x
7. Dorado-Garcia A, Smid JH, van Pelt W, et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J Antimicrob Chemother. 2018;73(2):339–347. doi:10.1093/jac/dkx397
8. Umeda K, Hase A, Matsuo M, Horimoto T, Ogasawara J. Prevalence and genetic characterization of cephalosporin-resistant Enterobacteriaceae among dogs and cats in an animal shelter. J Med Microbiol. 2019;68(3):339–345. doi:10.1099/jmm.0.000933
9. Hata A, Fujitani N, Ono F, Yoshikawa Y. Surveillance of antimicrobial-resistant Escherichia coli in Sheltered dogs in the Kanto Region of Japan. Sci Rep. 2022;12(1):773. doi:10.1038/s41598-021-04435-w
10. Madec JY, Haenni M, Nordmann P, Poirel L. Extended-spectrum beta-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans? Clin Microbiol Infect. 2017;23(11):826–833. doi:10.1016/j.cmi.2017.01.013
11. Bhat AH. Bacterial zoonoses transmitted by household pets and as reservoirs of antimicrobial resistant bacteria. Microb Pathog. 2021;155:104891. doi:10.1016/j.micpath.2021.104891
12. Marchetti L, Buldain D, Gortari Castillo L, Buchamer A, Chirino-Trejo M, Mestorino N. Pet and stray dogs as reservoirs of antimicrobial-resistant Escherichia coli. Int J Microbiol. 2021;2021:6664557. doi:10.1155/2021/6664557
13. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi:10.1089/cmb.2012.0021
14. Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom. 2018;4(7). doi:10.1099/mgen.0.000192
15. Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi:10.1186/s13059-014-0524-x
16. Rousham EK, Unicomb L, Islam MA. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc Biol Sci. 2018;285(1876). doi:10.1098/rspb.2018.0332
17. Booton RD, Meeyai A, Alhusein N, et al. One Health drivers of antibacterial resistance: quantifying the relative impacts of human, animal and environmental use and transmission. One Health. 2021;12:100220. doi:10.1016/j.onehlt.2021.100220
18. Costa D, Poeta P, Saenz Y, et al. Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet Microbiol. 2008;127(1–2):97–105. doi:10.1016/j.vetmic.2007.08.004
19. Wedley AL, Dawson S, Maddox TW, et al. Carriage of antimicrobial resistant Escherichia coli in dogs: prevalence, associated risk factors and molecular characteristics. Vet Microbiol. 2017;199:23–30. doi:10.1016/j.vetmic.2016.11.017
20. Katip W, Yoodee J, Uitrakul S, Oberdorfer P. Efficacy of loading dose colistin versus carbapenems for treatment of extended spectrum beta lactamase producing Enterobacteriaceae. Sci Rep. 2021;11(1):18. doi:10.1038/s41598-020-78098-4
21. De Graef EM, Decostere A, Devriese LA, Haesebrouck F. Antibiotic resistance among fecal indicator bacteria from healthy individually owned and kennel dogs. Microb Drug Resist. 2004;10(1):65–69. doi:10.1089/107662904323047826
22. Cozma AP, Rimbu CM, Zendri F, Maciuca IE, Timofte D. Clonal dissemination of extended-spectrum cephalosporin-resistant Enterobacterales between dogs and humans in households and animal shelters of Romania. Antibiotics. 2022;11(9):1242. doi:10.3390/antibiotics11091242
23. Masui T, Nakano R, Nakano A, et al. Predominance of CTX-M-9 group among ESBL-producing Escherichia coli isolated from healthy individuals in Japan. Microb Drug Resist. 2022;28(3):355–360. doi:10.1089/mdr.2021.0062
24. Rao L, Lv L, Zeng Z, et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet Microbiol. 2014;172(3–4):534–541. doi:10.1016/j.vetmic.2014.06.013
25. Lopez-Cerero L, Egea P, Serrano L, et al. Characterisation of clinical and food animal Escherichia coli isolates producing CTX-M-15 extended-spectrum beta-lactamase belonging to ST410 phylogroup A. Int J Antimicrob Agents. 2011;37(4):365–367. doi:10.1016/j.ijantimicag.2011.01.001
26. Xia S, Fan X, Huang Z, et al. Dominance of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS One. 2014;9(7):e100707. doi:10.1371/journal.pone.0100707
27. Freeman JT, Williamson DA, Heffernan H, Smith M, Bower JE, Roberts SA. Comparative epidemiology of CTX-M-14 and CTX-M-15 producing Escherichia coli: association with distinct demographic groups in the community in New Zealand. Eur J Clin Microbiol Infect Dis. 2012;31(8):2057–2060. doi:10.1007/s10096-011-1540-3
28. Maeyama Y, Taniguchi Y, Hayashi W, et al. Prevalence of ESBL/AmpC genes and specific clones among the third-generation cephalosporin-resistant Enterobacteriaceae from canine and feline clinical specimens in Japan. Vet Microbiol. 2018;216:183–189. doi:10.1016/j.vetmic.2018.02.020
29. Tamang MD, Nam HM, Jang GC, et al. Molecular characterization of extended-spectrum-beta-lactamase-producing and plasmid-mediated AmpC beta-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob Agents Chemother. 2012;56(5):2705–2712. doi:10.1128/AAC.05598-11
30. Peirano G, Bradford PA, Kazmierczak KM, et al. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis. 2014;20(11):1928–1931. doi:10.3201/eid2011.141388
31. Roer L, Overballe-Petersen S, Hansen F, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. 2018;3(4). doi:10.1128/mSphere.00337-18
32. Royer G, Darty MM, Clermont O, et al. Phylogroup stability contrasts with high within sequence type complex dynamics of Escherichia coli bloodstream infection isolates over a 12-year period. Genome Med. 2021;13(1):77. doi:10.1186/s13073-021-00892-0
33. Pitout JD, Laupland KB, Church DL, Menard ML, Johnson JR. Virulence factors of Escherichia coli isolates that produce CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2005;49(11):4667–4670. doi:10.1128/AAC.49.11.4667-4670.2005
34. Chattaway MA, Jenkins C, Ciesielczuk H, et al. Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg Infect Dis. 2014;20(11):1935–1937. doi:10.3201/eid2011.131845
35. Canton R, Akova M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18(5):413–431. doi:10.1111/j.1469-0691.2012.03821.x
36. Naseer U, The SA. CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist. 2011;17(1):83–97. doi:10.1089/mdr.2010.0132
37. Valverde A, Canton R, Garcillan-Barcia MP, et al. Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother. 2009;53(12):5204–5212. doi:10.1128/AAC.01706-08
38. Salgado-Caxito M, Benavides JA, Adell AD, Paes AC, Moreno-Switt AI. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats - A scoping review and meta-analysis. One Health. 2021;12:100236. doi:10.1016/j.onehlt.2021.100236
39. Bouchand C, Andréo A, Le Gallou F, et al. Retrospective analysis of a large single cohort of Enterobacteriaceae producing extended-spectrum B-lactamase (E-ESBL) patients: incidence, microbiology, and mortality. Eur J Clin Microbiol Infect Dis. 2022;41(10):1237–1243. doi:10.1007/s10096-022-04489-2
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

