Back to Journals » Infection and Drug Resistance » Volume 16
Genomic Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Isolated from Neonatal Patients in Southwest China During 2017–2021
Authors Wu W, Jiang Y, Zhou W, Kuang L
Received 5 July 2023
Accepted for publication 21 September 2023
Published 17 October 2023 Volume 2023:16 Pages 6725—6733
DOI https://doi.org/10.2147/IDR.S426565
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Wenjing Wu,1,2 Yongmei Jiang,1,2 Wei Zhou,1,2 Linghan Kuang1,2
1Department of Laboratory Medicine, West China Second University Hospital, Sichuan University, Chengdu, Sichuan, 610041, People’s Republic of China; 2Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, Sichuan, 610041, People’s Republic of China
Correspondence: Linghan Kuang, Department of Laboratory Medicine, West China Second University Hospital, Sichuan University, No. 20, Section 3, Renmin South Road, Chengdu, Sichuan, 610041, People’s Republic of China, Tel +86-28-8550-1201, Email [email protected]
Objective: Carbapenem-resistant Klebsiella pneumoniae (CRKP) is spreading worldwide, becoming a serious threat to public health. The present study aimed to analyze the molecular epidemiology and drug resistance mechanism of CRKP isolated from neonatal patients in Sichuan, Southwest China.
Methods: CRKP isolates were collected from neonatal patients of West China Second University Hospital from June 2017 to June 2021. Antimicrobial susceptibility testing was performed using broth microdilution. Whole-genome sequencing of all isolates were performed to determine the antimicrobial resistance genes, sequence typing, phylogenetic relationships.
Results: In total, 41 nonduplicate CRKP isolates were collected. All isolates were highly resistant to the cephalosporins and carbapenems, however, they were all susceptible to amikacin, tigecycline, ciprofloxacin, and colistin. Various resistance genes were detected, blaNDM-5 (n = 35, 85.4%) was the predominant carbapenemase genes. The most common replicon type was IncX3, which was harbored by 36 (87.8%) isolates, followed by IncFIB (n = 34, 82.9%), and IncFII (n = 32, 78.0%). The 41 CRKP isolates belonged to 8 sequence types (STs) and ST789 (n = 29, all had blaNDM-5) was the dominant sequence type.
Conclusion: The study revealed that blaNDM was the most dominant carbapenemase resistance gene. ST789 CRKP strains carrying blaNDM-5 were a tremendous menace to neonates in this hospital. Therefore, effectively implement prevention and control measures need to be taken for the prevention and treatment of CRKP infection in the neonatal ward.
Keywords: Klebsiella pneumoniae, carbapenemase, NDM-5, ST789, neonates
Introduction
Klebsiella pneumoniae (K. pneumoniae), which belongs to the family Enterobacteriaceae, is a common nosocomial pathogen that causes a variety of human infections, including bloodstream infection, respiratory tract infection, urinary tract infection, and abdominal infection.1,2 Carbapenems are generally considered to be a last choice for the treatment of multi-drug resistant gram-negative bacterial infections. However, with the large or unreasonable use of carbapenems antibiotics in recent years, carbapenem-resistant Klebsiella pneumoniae (CRKP) has emerged and become a serious threat to public health, which can cause higher morbidity and mortality.3,4
According to the data of WHO global antimicrobial resistance surveillance system (GLASS) in 2021,5 the resistance rates of meropenem and imipenem for CRKP were 12.34% and 10.63% respectively. Surveillance reports from China Antimicrobial Resistance Surveillance Network (CHINET) show that the meropenem resistance rate in a K. pneumoniae increased from 2.9% in 2005 to 24.4% in 2021.6 Data from the Infectious Disease Surveillance of Pediatrics (ISPED) program indicates that the proportion of carbapenem-resistant Enterobacteriaceae (CRE) in the neonatal group is much greater than in the non-neonatal group (11.1% vs 5.5%), and CRE has been presenting the potential threat to the health of children in China, especially to the immature immunity neonates.7 Therefore, more attention needs to be paid to the CRKP infections in neonate.
Carbapenem resistance in K. pneumoniae is mainly due to the following mechanisms: (1) the production of carbapenemases, including KPC (class A), NDM, VIM and IMP (class B), and OXA-48 (class D) enzymes, (2) loss of outer membrane proteins and (3) overexpression of efflux pumps.8 Carbapenemase production is the main resistance mechanism. The dissemination of CRKP is clonal, and the population structure is geographically specific. It has been reported that sequence type (ST) 258 has become the most predominant type in the United States, and Europe9 and ST11 is the predominant type in China.10 The bacterial epidemiology and resistance profiles in children are quite different from adult population in China. blaKPC-2 was the most prevalent carbapenemase gene in K. pneumoniae isolated from Chinese adults, and blaNDM was the most dominant gene among children.11,12 In addition, the resistance and prevalence of CRKP in different regions of China are also different.13 However, the molecular characteristics of CRKP in neonate are largely lacking or very limited. On this basis, the study collected CRKP isolates from neonatal patients of West China Second University Hospital in Sichuan, Southwest China, and used whole-genome sequencing (WGS) technology to analyze the molecular epidemiology and drug resistance mechanism of neonatal CRKP to provide evidence for clinical treatment and effective control of nosocomial infection with CRKP.
Materials and Methods
Bacterial Isolates, Identification, and Susceptibility Testing
In this study, nonduplicate CRKP isolates were collected from neonatal patients hospitalized in the neonatal intensive care unit (NICU) and the general neonatal wards of West China Second University Hospital from June 2017 to June 2021. West China Second University Hospital is a tertiary-care teaching hospital located in Sichuan, Southwest China, where over 6000 neonates are hospitalized every year. All isolates were initially identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, bioMérieux, France). Antimicrobial susceptibility testing was performed using broth microdilution with the Vitek II automated system (bioMérieux). In addition, minimum inhibitory concentrations (MICs) of colistin and ceftazidime-avibactam against the isolates were determined using the broth microdilution according to the Clinical Laboratory Standards Institute (CLSI). The results were interpreted according to the criteria of CLSI 2022 breakpoints. For tigecycline and colistin, which were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria (https://www.eucast.org). Escherichia coli ATCC 25922 was used as a quality control.
String Testing
The hypermucoviscous phenotype was detected by string test. All isolates were cultured at 35°C for overnight on Columbia blood agar medium. The formation of viscous string >5 mm in length was considered positive for the string test when the bacterial colonies were stretched by an inoculation loop.14
Genomic DNA Extraction, Whole-Genome Sequencing, Assembly, and Annotation
Genomic DNA was prepared using the QIAamp DNA mini kit (Qiagen, Hilden, Germany), and sequencing libraries were generated using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following the manufacturer’s recommendations. Whole genome sequencing was carried out using the HiSeq X10 platform (Illumina, San Diego, CA, USA). Reads were trimmed using Trimmomatic v0.3915 under the default setting and were de novo assembled into contigs using SPAdes v3.14.0.16 Prokka v1.14.517 was used to annotate the assembled genomes.
Genome Profiling
Antimicrobial resistance genes (ARGs) and plasmid replicons were identified using ResFinder and PlasmidFinder (http://genomicepidemiology.org/). Sequence types (STs) were determined using the genome sequence to query the multilocus sequence typing database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Capsule typing and virulence factor prediction were performed using Kleborate v2.2.0 (https://github.com/katholt/Kleborate).
Phylogenetic Analysis
We performed single-nucleotide polymorphism (SNP) calling for genome sequences to untangle the clonal relatedness of strains and to investigate whether the ST789 strains in this study are clonally related to strains recovered elsewhere. We used the complete genome of strain 090573 (GenBank accession no. CP071167), an ST789 recovered from sputum in China in 2019, as the reference sequence for mapping. The quality-controlled reads of each strain were mapped against the reference using Snippy version 4.3.6 (https://github.com/tseemann/snippy) with default settings. A pseudo-multiple-alignment was generated using the bundled script and the phylogeny was then inferred using RaxML v8.2.1218 under GTRGAMMA model with a 1000-bootstrap test. Phylogenetic trees were viewed in Figtree v1.4.4 (https://github.com/rambaut/figtree/) and annotated using iTOL v6.7.5.19
Nucleotide Sequence Accession Numbers
The draft genomes of strains have been deposited in GenBank under the BioProject number PRJNA847410.
Results
Clinical Characteristics of CRKP Isolates
In total, 41 nonduplicate CRKP isolates were collected from June 2017 to June 2021, the male-to-female ratio was 1:1.05, the median age was 19 days (interquartile range: 13–31 days). The CRKP isolates were obtained from sputum (39.0%, n = 16), blood (26.8%, n = 11), tracheal tube (19.5%, n = 8), pleural and peritoneal effusions (7.3%, n = 3), secretions (4.9%, n = 2), and urine (2.4%, n = 1) (Figure S1). The majority of patients were born prematurely (75.6%, 31/41), 65.9% (27/41) were born by caesarean section, 70.7% (29/41) had received mechanical ventilation, and 63.4% (26/41) had peripherally inserted central catheter placed. Further demographic and clinical details are provided in Table 1.
 |
Table 1 Clinical Characteristics of CRKP Patients |
Antimicrobial Susceptibility Testing and Hypermucoviscous Phenotype
The 41 CRKP strains demonstrated high resistance to all β-lactams and their corresponding inhibitors, including cefuroxime, ceftazidime, ceftriaxone, cefepime, cefoperazone/sulbactam, ceftazidime-avibactam, piperacillin/tazobactam, imipenem, meropenem, and ertapenem. Various levels of susceptibilities to aztreonam (24.4%, 10/41), sulfamethoxazole-trimethoprim (90.2%, 37/41), tobramycin (92.7%, 38/41), gentamicin (95.1%, 39/41), and levofloxacin (95.1%, 39/41) were observed. Moreover, all the CRKP isolates (100%) were susceptible to amikacin, tigecycline, ciprofloxacin, and colistin (Table 2 and Table S1). None of the 41 isolates were hypermucoviscous.
 |
Table 2 Antimicrobial Susceptibility and MIC Distributions of CRKP Strains |
Distribution of Antimicrobial Resistance Genes and Virulence Genes
Various resistance genes were identified in the 41 CRKP isolates (Figure1), 39 strains were found to carry carbapenemase genes, among which blaNDM-5 was the predominant one (n = 35, 85.4%), followed by blaNDM-1 (n = 3, 7.3%), and blaIMP-4 (n = 1, 2.4%). However, two strains did not carry any carbapenemase gene. blaSHV (n = 41, 100%), blaCTX-M (n = 35, 83.3%) and blaTEM-1 (n =30, 97.6%) also had high detection rates. The AmpC beta-lactamase, blaDHA-1 (n = 5, 11.9%) was identified. In addition, the fosfomycin resistance gene fosA and tetracycline resistance gene tet (34) were detected in all isolates (Table S2). By analyzing virulence genes, none of the CRKP isolates carried the rmpA/rmpA2 gene (encoding mucoid phenotype regulators), iucABCD–iutA (encoding aerobactin), and iroBCDN (encoding salmochelin). The genes encoding yersiniabactin (irp/ybtAEPQSTUX) were detected in 5 isolates, which all belonged to ST2407.
 |
Figure 1 The phylogenetic tree and the distribution of antibiotic resistance genes of the 41 CRKP isolates. |
STs, Capsular Wzi Types and Plasmid Replicons of the 42 CRKP Strains
MLST results showed that a total of 8 different STs were identified for the 41 CRKP isolates. ST789 (70.7%, 29/42) was the most prevalent, followed by ST2407 (12.2%, 5/42) and ST17 (4.9%, 2/42). Other STs, including ST29, ST35, ST70, ST278, and ST307 were only detected in one isolate (Figure 2). All 29 ST789 isolates carried blaNDM-5, blaCTX-M-15, blaSHV-25, and blaTEM-1. All 5 ST2407 isolates carried blaNDM-5, blaCTX-M-14, blaSHV-187 and blaDHA-1. Four serotypes were identified in the 41 CRKP isolates, KL18 (n = 29, 70.7%) was the most common one, followed by KL25 (n = 5, 12.2%), KL22 (n = 1, 2.4%) and KL30 (n = 1, 2.4%). In addition, five isolates were unclassified. Seven capsular wzi types were identified, wzi18 (n = 29, 70.7%) was the most common one, which all isolates belonged to ST789 (Table S2). Eleven types of plasmid replicons were identified in the 41 isolates. The most common replicon type was IncX3, which was harbored by 36 (87.8%) isolates, followed by IncFIB (n = 34, 82.9%), and IncFII (n = 32, 78.0%). The remaining plasmid replicons, such as Col(pHAD28) and IncR, were relatively less common (Figure 3). Among the isolates, the most common type of replicon combination was IncX3 and IncFIB, accounting for 75.6% (31/41) (Table S2).
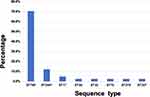 |
Figure 2 Sequence types of the 41 CRKP isolates. |
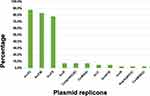 |
Figure 3 Plasmid replicons among the 41 CRKP isolates. |
Discussion
The emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) has been regarded as a worldwide public health concern, and the epidemic characteristics of CRKP vary by region. The prevalence of CRE in the children is increasing, and the neonatal intensive care unit (NICU) patients had the highest proportion of CRKP nosocomial infections, who showed different molecular and epidemiological characteristics of CRKP from non-neonatal patients.20 Therefore, it is necessary to understand the clinical and molecular characteristics of CRKP strains in neonates. In this study, we used whole-genome sequencing technology to analyze the genomic characteristics of neonatal CRKP strains collected in a tertiary hospital in Southwest China. The 41 CRKP isolates collected in this study were mainly from sputum (39%) which might be associated with respiratory insufficiency and mechanical ventilation. It had been reported that many risk factors, including mechanical ventilation, cesarean section, prior exposure to broad-spectrum beta-lactams, the use of central venous catheter, and low birth weight were associated with CRKP infection and/or colonization in pediatric.21–24 In this study, the great majority of neonates shared similar clinical features with premature birth (75.6%), caesarean birth (65.9%), venous catheterization (70.7%), and mechanical ventilation (70.7%). More than half of the healthcare-associated infections were occurred in patients of ≤1000 g weight at birth in NICU.25 Therefore, greater attention should be paid to neonates who are of low weight and undergoing mechanical ventilation and invasive procedures, to reduce the incidence of CRKP infection.
The results of drug sensitivity test showed that all the CRKP strains were highly resistant to antibiotics widely used in children, such as cephalosporins, carbapenems, monobactams, etc, and had a low resistance to quinolones, sulfonamides, aminoglycoside antibiotics, which were consistent with the results of previous studies.26 However, other research reported that the resistance rates of CRKP to levofloxaxin, gentamicin and trimethoprim/sulfamethoxazole showed an increasing trend, suggesting the importance of dynamic monitoring.27,28 In addition, most strains showed high resistance rate to the new cephalosporin/beta-lactamase — ceftazidime-avibactam (95.1%), which was regarded as an effective drug for the treatment of CRKP infections, as compared with previously developed antimicrobials such as colistin, aminoglycosides or tigecycline.29,30 Due to the limited availability of effective drugs, the treatment of CRKP infections in children, particularly in newborns, presents a significant challenge. Combination therapy utilizing antibiotics with diverse mechanisms of action, such as polymyxin, aminoglycosides, fosfomycin, and carbapenems, has shown promising results in adult patients. However, there is a scarcity of data regarding the efficacy of these treatments in the pediatric population. Some studies have indicated that colistin may be an effective and safe option for treating neonatal infections caused by carbapenem-resistant Enterobacteriaceae (CRE).31–33 Previous studies have shown that fosfomycin-containing regimens, whether in combination with a carbapenem or not, appear to be effective for treating CRKP infections in neonates, offering a new avenue for therapy.34 In this study, it was observed that the strains were sensitive to certain antibiotics in vitro, suggesting that these drugs can potentially be utilized for neonatal CRKP infections in our hospital. Overall, the treatment of CRKP infections in children presents as a complex and challenging task. Consequently, antimicrobial therapy should be individualized, considering various factors including the severity of the patient’s illness, epidemiological characteristics, the source of infection, and the susceptibility profile of the isolated bacteria.
The production of carbapenemase is the major mechanism of carbapenem resistance in K. pneumoniae. It has been reported that NDM was the most common carbapenemase type in neonatal CRKP, followed by KPC and OXA-48-like.35 NDM belongs to the type of metallo-β-lactamase (MBL) that can hydrolyze most β-lactams (including carbapenems) but not monobactams, and cannot be inhibited by β-lactamase inhibitors, such as avibactam, clavulanic acid, sulbactam, and tazobactam, but can be inhibited by the MBL inhibitor EDTA.36
NDM-1 and NDM-5 are the most common NDM variants in CRKP and carbapenem-resistant Escherichia coli respectively, and NDM-5 showed higher hydrolytic stability for imipenem and meropenem as compared to NDM-1.8 In this study, 38 strains of CRKP carried blaNDM, among which 35 strains carried blaNDM-5 and 3 strains carried blaNDM-1, which interpreted the high resistance rate to ceftazidime-avibactam of the strains. In addition, blakpc was not detected, which was the most common in adults. It indicated that the therapeutic regimen of CRKP would be different between children and adults. In contrast to the previous finding that blaNDM-1 was the most prevalent in neonatal patients, we found that blaNDM-5 was the most prevalent type in this study.34 NDM-5-producing K. pneumoniae in neonates has been reported in China, India, Nigeria and Vietnam.35 The outbreak of blaNDM-5-carrying ST337 K. pneumoniae in the neonatal ward was first reported in Jiangsu, and all the blaNDM-5 gene located on the IncX3 type plasmid.37 Moreover, high prevalence of the combination of the β-lactamases genes of blaCTX-M, blaSHV, and blaTEM among the NDM-producing strains was observed in this study, and the coexistence of drug-resistant determinants with the combination of carbapenemase and two or more β-lactamases leaded to the multidrug resistance. Two isolates in the study did not carry any carbapenemase genes, and their MICs of carbapenem were not high (4–8 μg/mL). The mechanism of carbapenem resistance remains to be studied in the future. Both phenotypic and genomic analysis showed that the CRKP isolates were not hypermucoviscous, which was highly correlated with hypervirulent Klebsiella pneumoniae, suggesting that the CRKP strains in our hospital were not yet highly lethal and virulent.
In this study, the CRKP with carbapenemases belonged to different STs, ST789, ST2407, ST17, ST29, ST35, and ST278, implying the genetic diversity of CRKP isolates. ST11 is the dominant ST in Asia for CRKP, which accounts for up to 60% of CRKP in China.10 However, ST789 was identified as the most prevalent type, and no ST11 CRKP was isolated in our study. ST789 is not a common type, and KPC-2-producing Klebsiella pneumoniae ST789 has been reported in some countries, such as China, Malaysia, South Korea, and the USA.38 In addition, previous studies reported that blaNDM-5-carring ST789 CRKP caused multiple outbreaks involved in 2 neonatal ICUs at different hospitals in Sichuan.38,39 Then NDM-5-producing ST789 CRKP may become an emerging threat for neonates in this region and need rigorous monitoring. ST2407 was the second most common sequence type, and it had been reported that ST2407 was the dominant type of blaNDM-carrying Klebsiella pneumoniae in one hospital from Chengdu and clone transmission occurred in this hospital.40 Plasmids have the ability to carry multiple antibiotic resistance genes and can transfer between the same or different species by conjugation, which makes them play an important role in the dissemination of antimicrobial resistance.41 In our study, eleven types of plasmid replicons were identified in the 41 isolates, among them IncX3 was the most common plasmid type. It suggested that IncX3 plasmids may be a major vehicle in mediating the dissemination of blaNDM in East Asia, particularly in China.42 Key plasmids (IncX3) and ST diversity accelerated the spread of blaNDM in CRKP.40
There are also some limitations in our study. It was performed in a single tertiary hospital in Sichuan, and the results cannot be generalized to other institutions. Multi-center isolates should be collected to provide more evidence on the clinical significance of CRKP strains.
Conclusion
The study revealed the prevalence of CRKP strains isolated from neonatal patients. BlaNDM was the most dominant carbapenemase resistance gene. ST789 CRKP strains carrying blaNDM-5 were a tremendous menace to neonates in this hospital. Therefore, effectively implement prevention and control measures need to be taken for the prevention and treatment of CRKP infection in the neonatal ward.
Ethical Approval
The study was approved by Clinical Trial Ethics Committee of West China Second University Hospital, Sichuan University (2021-119). The present study was a retrospective study focusing on bacteria and did not contain any sensitive personal information. Therefore, informed consent was not required in line with local legislation.
Funding
The work was supported by the Sichuan Science and Technology Program (No. 2022NSFSC1420 to WJW and No. 2023NSFSC1699 to LHK).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Diekema DJ, Hsueh PR, Mendes RE, et al. The microbiology of bloodstream infection: 20-year trends from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother. 2019;63(7). doi:10.1128/AAC.00355-19
2. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
3. Stewardson AJ, Marimuthu K, Sengupta S, et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis. 2019;19(6):601–610. doi:10.1016/S1473-3099(18)30792-8
4. Zhou R, Fang X, Zhang J, et al. Impact of carbapenem resistance on mortality in patients infected with Enterobacteriaceae: a systematic review and meta-analysis. BMJ Open. 2021;11(12):e054971. doi:10.1136/bmjopen-2021-054971
5. World Health Organization. Global Antimicrobial Resistance and use Surveillance System (GLASS) Report 2021. Geneva: World Health Organization; 2021.
6. Fupin HU, Yan GUO, Demei ZHU, et al. CHINET surveillance of antimicrobial resistance among the bacterial isolates in 2021. Chin J Infect Chemother. 2022;22(5):521–530. doi:10.16718/j.1009-7708.2022.05.001
7. Fu P, Xu H, Jing C, et al. Bacterial epidemiology and antimicrobial resistance profiles in children reported by the ISPED program in China, 2016 to 2020. Microbiol Spectr. 2021;9(3):e0028321. doi:10.1128/Spectrum.00283-21
8. Ma J, Song X, Li M, et al. Global spread of carbapenem-resistant Enterobacteriaceae: epidemiological features, resistance mechanisms, detection and therapy. Microbiol Res. 2023;266:127249. doi:10.1016/j.micres.2022.127249
9. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi:10.1128/AAC.01019-15
10. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
11. Gao F, Xiong Z, Liang B, et al. Molecular characterization and epidemiology of carbapenem-resistant Enterobacteriaceae isolated from pediatric patients in Guangzhou, Southern China. Can J Infect Dis Med Microbiol. 2023;2023:4762143. doi:10.1155/2023/4762143
12. Han R, Shi Q, Wu S, et al. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
13. Tao G, Tan H, Ma J, Chen Q. Resistance phenotype and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae isolated from Nanjing children’s hospital in Jiangsu Province, China. Infect Drug Resist. 2022;15:5435–5447. doi:10.2147/IDR.S377068
14. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi:10.4161/viru.22718
15. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170
16. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi:10.1089/cmb.2012.0021
17. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
18. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi:10.1093/bioinformatics/btu033
19. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi:10.1093/nar/gkab301
20. Yin L, He L, Miao J, et al. Actively surveillance and appropriate patients placements’ contact isolation dramatically decreased Carbapenem-Resistant Enterobacteriaceae infection and colonization in pediatric patients in China. J Hosp Infect. 2020;105(3):486–494. doi:10.1016/j.jhin.2020.03.031
21. Wang J, Lv Y, Yang W, Zhao P, Yin C. Epidemiology and clinical characteristics of infection/colonization due to carbapenemase-producing Enterobacterales in neonatal patients. BMC Microbiol. 2022;22(1):177. doi:10.1186/s12866-022-02585-z
22. Darda VM, Iosifidis E, Antachopoulos C, et al. Modifiable risk factors associated with later gut decolonization of carbapenem-resistant Gram-negative bacteria in children: a prospective cohort study. J Hosp Infect. 2023;136:75–84. doi:10.1016/j.jhin.2023.03.024
23. Yen CS, Hsiao HL, Lee CC, et al. Carbapenem-resistant Enterobacteriaceae infection in children less than one year old in an Asian medical center. Pediatr Neonatol. 2023;64(2):168–175. doi:10.1016/j.pedneo.2022.05.016
24. Yin L, He L, Miao J, et al. Carbapenem-resistant Enterobacterales colonization and subsequent infection in a neonatal intensive care unit in Shanghai, China. Infect Prev Pract. 2021;3(3):100147. doi:10.1016/j.infpip.2021.100147
25. Scamardo MS, Dolce P, Esposito EP, Raimondi F, Triassi M, Zarrilli R. Trends, risk factors and outcomes of healthcare-associated infections in a neonatal intensive care unit in Italy during 2013–2017. Ital J Pediatr. 2020;46(1):34. doi:10.1186/s13052-020-0799-3
26. Zhang X, Xue J, Shen MJ, An WH, Chen ZQ, Wu KF. Molecular typing and drug resistance analysis of carbapenem-resistant Klebsiella pneumoniae from paediatric patients in China. J Infect Dev Ctries. 2022;16(11):1726–1731. doi:10.3855/jidc.17003
27. Yin L, Yan G, Lu L, et al. Molecular characteristics and virulence factors of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai, China. Infect Genet Evol. 2023;112:105451. doi:10.1016/j.meegid.2023.105451
28. Fu P, Luo X, Shen J, et al. The molecular and epidemiological characteristics of carbapenemase-producing Enterobacteriaceae isolated from children in Shanghai, China, 2016–2021. J Microbiol Immunol Infect. 2023;56(1):48–56. doi:10.1016/j.jmii.2022.07.012
29. Shields RK, Nguyen MH, Chen L, et al. Ceftazidime-avibactam is superior to other treatment regimens against carbapenem-resistant Klebsiella pneumoniae bacteremia. Antimicrob Agents Chemother. 2017;61(8). doi:10.1128/AAC.00883-17
30. Carmeli Y, Armstrong J, Laud PJ, et al. Ceftazidime-avibactam or best available therapy in patients with ceftazidime-resistant Enterobacteriaceae and Pseudomonas aeruginosa complicated urinary tract infections or complicated intra-abdominal infections (REPRISE): a randomised, pathogen-directed, Phase 3 study. Lancet Infect Dis. 2016;16(6):661–673. doi:10.1016/S1473-3099(16)30004-4
31. Ipek MS, Aktar F, Okur N, Celik M, Ozbek E. Colistin use in critically ill neonates: a case-control study. Pediatr Neonatol. 2017;58(6):490–496. doi:10.1016/j.pedneo.2016.10.002
32. Cagan E, Kiray Bas E, Asker HS. Use of colistin in a neonatal intensive care unit: a cohort study of 65 patients. Med Sci Monit. 2017;23:548–554. doi:10.12659/msm.898213
33. Abrahams I, Dramowski A, Moloto K, Lloyd L, Whitelaw A, Bekker A. Colistin use in a carbapenem-resistant Enterobacterales outbreak at a South African neonatal unit. S Afr J Infect Dis. 2023;38(1):487. doi:10.4102/sajid.v38i1.487
34. Yin D, Zhang L, Wang A, et al. Clinical and molecular epidemiologic characteristics of carbapenem-resistant Klebsiella pneumoniae infection/colonization among neonates in China. J Hosp Infect. 2018;100(1):21–28. doi:10.1016/j.jhin.2018.05.005
35. Hu Y, Yang Y, Feng Y, et al. Prevalence and clonal diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal infections: a systematic review of 128 articles across 30 countries. PLoS Med. 2023;20(6):e1004233. doi:10.1371/journal.pmed.1004233
36. Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of new delhi metallo-beta-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17(1):101. doi:10.1186/s12866-017-1012-8
37. Kong Z, Cai R, Cheng C, et al. First reported nosocomial outbreak of NDM-5-Producing Klebsiella pneumoniae In A neonatal unit in China. Infect Drug Resist. 2019;12:3557–3566. doi:10.2147/IDR.S218945
38. Qiao F, Wei L, Feng Y, et al. Handwashing sink contamination and carbapenem-resistant Klebsiella infection in the intensive care unit: a prospective multicenter study. Clin Infect Dis. 2020;71(Suppl 4):S379–S385. doi:10.1093/cid/ciaa1515
39. Wei L, Feng Y, Wen H, Ya H, Qiao F, Zong Z. NDM-5-producing carbapenem-resistant Klebsiella pneumoniae of sequence type 789 emerged as a threat for neonates: a multicentre, genome-based study. Int J Antimicrob Agents. 2022;59(2):106508. doi:10.1016/j.ijantimicag.2021.106508
40. Fu B, Yin D, Sun C, et al. Clonal and Horizontal Transmission of bla(NDM) among Klebsiella pneumoniae in Children’s Intensive Care Units. Microbiol Spectr. 2022;10(4):e0157421. doi:10.1128/spectrum.01574-21
41. Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann N Y Acad Sci. 2019;1457(1):61–91. doi:10.1111/nyas.14223
42. Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32(2). doi:10.1128/CMR.00115-18
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
