Back to Journals » Infection and Drug Resistance » Volume 14
Genetic Variability in Patients with HCV-Related Hepatocellular Carcinoma
Authors Pisaturo M, Di Fraia A, Occhiello L , Minichini C, Starace M , Iodice V, Farella N, Stanzione M, Coppola N
Received 10 September 2021
Accepted for publication 9 November 2021
Published 7 December 2021 Volume 2021:14 Pages 5199—5208
DOI https://doi.org/10.2147/IDR.S337647
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Mariantonietta Pisaturo,1 Alessandra Di Fraia,1 Laura Occhiello,1 Carmine Minichini,1 Mario Starace,1 Valentina Iodice,2 Nunzia Farella,2 Maria Stanzione,1 Nicola Coppola1
1Infectious Diseases, Department of Mental Health and Public Medicine, University of Campania “L. Vanvitelli”, Naples, Italy; 2IX Infectious Disease Unit, AORN dei Colli, Naples, Italy
Correspondence: Nicola Coppola
Department of Mental Health and Public Medicine, Section of Infectious Diseases, University of Campania Luigi Vanvitelli, Via L. Armanni 5, Naples, 80131, Italy
Tel +39 081 5666223
Fax +39 081 5666013
Email [email protected]
Background: The present paper evaluates the genetic variability of HCV in patients with hepatocellular carcinoma (HCC).
Methods: Amino acid substitutions (aas) in NS3, NS5A and core regions were analyzed in 17 patients with HCC (Cases) and 13 without HCC (Controls), all naïve to DAAs. For the Cases, a sample of neoplastic liver tissue, non-neoplastic liver tissue and a serum sample were collected; for the Controls, a sample of liver tissue was collected. Sanger sequencing of three regions was performed using homemade protocols.
Results: Phylogenetic trees showed that there was no difference in the virus populations in the three compartments analyzed for the three HCV regions in patients with HCC. Low variability and no difference between the Cases and Controls were observed in the core and NS5A regions; however, in the NS3 region, a higher variability was observed in the Cases. No difference was observed in the core region between Cases and Controls. In NS3, aa substitutions at positions 103 and 122 were more frequently found in Cases than Controls (in both cases 50% vs 9.1%, p< 0.05); moreover, aas in positions 32, 44 (p=0.035 for both), 79 (p=0.008) and 121 (p=0.018) were observed in the Cases and absent in the Controls. Finally, considering the NS5A region, aa substitutions at positions 37 and 54 were more frequently identified in the Cases than the Controls, but without statistical significance.
Conclusion: These data may suggest a higher aa variability in patients with HCC than in those without, especially in the NS3 region.
Keywords: hepatocellular carcinoma, HCV, viral variability, chronic hepatitis C, chronic infection C, core region, NS5A region, NS3 region, aa substitutions, phylogenesis
Introduction
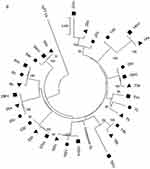
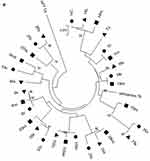
Hepatocellular carcinoma (HCC) is the sixth most frequent cancer worldwide and the fourth leading cause of cancer-related deaths and arises especially in cirrhotic patients.1 Figure 1 Continued. Figure 1 Continued.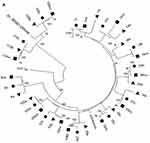
The prevalence of HCC reflects the epidemiology of the etiological agents responsible for chronic liver diseases. In developing countries, liver cancer is associated with hepatitis B virus (HBV) and hepatitis C virus (HCV) chronic infection. In developed countries, the impact of HBV and HCV on the epidemiology of HCC shows a reduction with an increasing role of non-alcoholic fatty liver diseases (NAFLD).2 In fact, the universal HBV vaccination has significantly reduced the incidence of HBV-related HCC, and the increasing use in recent years of directly acting antiviral agents significantly reduced, although perhaps not fully eliminate, the risk of developing HCC.3,4
Hepatocarcinogenesis is a complex multistep process and is influenced by many risk factors, including the oncogenic effect of HBV proteins, steatosis and insulin resistance, chronic inflammation and fibrosis, oxidative stress and chromosomal instability.5,6
HCV is indirectly involved in hepatocarcinogenesis through the development of cirrhosis: HCV-related necrosis may promote cell proliferation and long-term hepatic inflammation that may contribute to the development of HCC with oxidative stress and DNA damage.7–9 However, recent reports hypothesized that HCV directly favored the development of HCC by different molecular mechanisms, such as the alteration of normal cellular signaling pathways to stimulate host cell growth and cell transformation. Moreover, some studies have recently shown that some amino acid substitutions (aas) in the core region of genotype 1b were associated with HCC; specifically, aas 70 and/or 91 seem to be predictors of hepatocarcinogenesis.10,11
The present study evaluated the genetic variability of HCV in patients with HCC compared to those without by analyzing NS3, NS5A and core HCV regions in different specimens, serum, cancer liver tissue and non-cancer liver tissue.
Patients and Methods
Patients
Of the 68 consecutive patients who underwent a diagnostic liver biopsy for HCC at the liver unit of University of Campania, Naples, Southern Italy, from June 2013 to December 2014,12,13 17 HCV-RNA positive were retrospectively enrolled in the present study according to the availability of liver tissue collected (Case group). HCC was diagnosed according to the EASL/EORTC criteria.14 To reach the aim of the study, the first 13 patients without HCC who underwent liver biopsy to stage their liver disease in June–July 2013 and for whom the liver specimens collected at −80°C were available were enrolled (Control group).
A physical examination, full liver function tests and serology for HBsAg, anti-HCV and anti-HIV were performed in each patient enrolled. The stage of HCC was assessed according to the BCLC (Barcelona Clinic Liver Cancer) criteria.15
For the 17 Cases, a sample of cancer tissue (CT), non-neoplastic liver tissue (non-CT) and a serum sample (S) were collected for the aims of the study. For the 13 Controls, a sample of liver tissue was collected. Both for the Cases and Controls, fragments of nearly 3 mg were cut away from the two extremities of the liver biopsies and stored at −80°C in RNAlater solution (Qiagen GmbH, Hilden, Germany) for molecular analyses.
The study was approved by the Ethics Committee of the Azienda Ospedaliera Universitaria of the Second University of Naples (n°349/2013). All procedures performed in this study were in accordance with the ethics standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethics standards. Informed consent was obtained from all participants included in the study.
Routine Methods
Anti-HCV was performed using a third-generation commercial immunoenzymatic assay (Ortho Diagnostic Systems, Neckargemund, Germany). Liver biochemistry and routine analyses were performed by routine methods in a Cobas Modular 6000 automated analyzer using c501 biochemistry modules (Roche Diagnostics Ltd, Rotkreuz, Switzerland).
HCV Molecular Methods
Viral RNA was extracted from 200 μL of serum samples and from HCC (CT) and non-HCC (non-CT) tissues using a microspin column (QIAamp RNA viral kit and QIAamp DNA/RNA mini-kit, respectively, Qiagen GmbH, Hilden, Germany). Serum HCV‐RNA was quantified by a real-time PCR method in a Light cycler 1.5 (Roche Diagnostics, Branchburg, NJ, USA) with a sensibility of less than 40 IU/mL, as previously reported.16
HCV genotypes were identified by HCV genotype Lipa assay (Bayer, France).
Sanger sequencing of NS3, NS5A and core regions was performed on samples applying home‐made protocols.17 Briefly, the NS3 (181aa), NS5A (140aa) and Core (100 aa) regions were amplified by primers as shown in Supplementary Table 1 and sequenced using ABI 3500 Genetic Analyzer (Applied Biosystems).
The sequences were compared with reference sequences recognized in the literature and taken from Los Alamos Database and other databases (GenBank-accession-numbers: HCV-1a: H77 NC_004102, HCV-1b: D90208, HCV-2a/2c: D50409, HCV-3a: D28917, HCV-3h: JF735126, HCV-4: DQ418786). The phylogenetic trees were made using the Mega 10 program. By the comparison with the reference sequence, it was possible to establish the genotypes and possible contaminations of the samples. The mutations and quasi-species were identified by seqscape program software (Applied Biosystems) with a >20% tolerance for improper sequencing. Finally, we interpreted the amino acid (aa) substitutions based on the reference sources.
Statistical Analysis
Continuous variables were summarized as mean and standard deviation, and categorical variables as absolute and relative frequencies. For continuous variables, the differences were evaluated by the Student’s t-test; categorical variables were compared by the chi-square test, using exact procedures if needed. Differences were considered statistically significant at p<0.05.
Results
The demographic and biochemical characteristics of Cases and Controls are shown in Table 1. The Cases were older (median age 71 years, range 66–79) than the Controls (median age 60, range 42–71), (<0.00001) and mostly males (58.8% vs 46.2, a difference not significant to the statistical analysis.) (Table 1). Compared with the Controls, the Cases showed lower transaminase serum values [ALT, mean × normal value ± standard deviation (SD): 1.51± 0.99 vs 3.052±1.58, p=0.16; AST: 1.59± 0.97 vs 1.87± 1.07, p=0.71] (Table 1). On the contrary, total bilirubin value was higher in the Cases than Controls (1.64±1.17 vs 0.99± 0.50 mg/dL, p=0.10.) (Table 1). The serum HCV-RNA viral load was higher in the Controls (Table 1). The most representative HCV genotype was 1b in both groups: 14 (82%) patients in the Cases and 11 (85%) in the Controls; 3 Cases harbored HCV genotype 2c and 2 Controls genotype 2a (Table 1). All patients were naïve to DAAs.
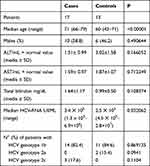 |
Table 1 Demographic and Clinical Characteristics in the Cases and Controls |
The sequence analyzed in the different HCV regions, core, NS3 and NS5A, for the Cases and Controls are shown in Supplementary Tables 2 and 3, respectively. The sequences of patients with HCC (Case group) obtained in the three compartments (cancer tissue, non-cancer tissue and serum) were used to create phylogenetic trees (Figure 1A–C). Phylogenetic trees showed that there was no difference between the virus populations in the three compartments analyzed in the patients with HCC (Figure 1).
Table 2 shows the HCV variability in the three viral regions analyzed in the Cases and Controls. Low variability and no difference between the Cases and the Controls were observed in the core and NS5A regions; however, in the NS3 region a higher variability was observed in the Cases. In fact, compared with the patients without, those with HCC showed in all three compartments a higher number of aa substitutions in the NS3 region: median (IQR) aa substitutions, 9 (9.5) in cancer tissue, 7.5 (12.75) in non-cancer tissue and 8 (24) in serum in the Cases vs 5 (4) in liver tissue in the Controls.
 |
Table 2 Number of aa Substitutions in Core, NS3 and NS5A Regions for the Cases and Controls |
Tables 3–5 show, for each HCV region analyzed, the positions in which there was more frequent amino acid substitutions (in at least 25% of the subjects).
 |
Table 3 Prevalence of Patients with More Frequent* Amino Acid Substitutions at Different Positions of the Core Region |
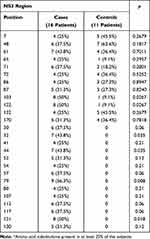 |
Table 4 Prevalence of Patients with More Frequent* Amino Acid Substitutions at Different Positions of the NS3 Region |
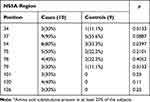 |
Table 5 Prevalence of Patients with More Frequent* Amino Acid Substitutions at Different Positions of the NS5A Region |
Table 6 shows the positions of amino acid (aa) substitutions in NS3, NS5A and core region, amino acid substitutions identified and the compartments in which these were found.
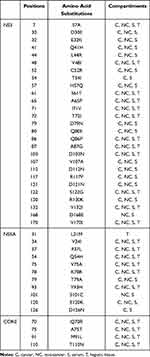 |
Table 6 Positions of Amino Acid (aa) Substitutions in NS3, NS5A and Core Region, Amino Acid Substitutions Identified and the Compartments in Which These Were Found |
Considering the HCV core region, no differences in the aa substitutions were found in the Cases and Controls (Table 3).
Considering the NS3 region, aa substitutions at positions 103 and 122 were more frequently found in the Cases than the Controls (in both cases 50% vs 9.1%, p<0.05; Table 4). Amino acid substitutions in positions 30, 32, 41, 44, 52, 54, 57, 79, 80, 107, 112, 117, 121 and 130 were also observed in the Cases, but were absent in the Controls, with a difference significant to the statistical analysis for position 32 and 44 (p=0.035 for both), for position 79 (p=0.008) and 121 (p=0.018) (Table 4). Interestingly, 2 aa substitutions (S122G and D168E) conferring resistance to anti-NS3 inhibitors were identified: 4 Cases and 1 Control harbored S122G and 1 Case D168E (Table 4).
Finally, considering the NS5A region, aa substitutions at position 37 and 54 were more frequently identified in the Cases than in the Controls, but due to the small number of patients, without statistical significance (90% vs 55.6% and 80% vs 33%, respectively, Table 5). Moreover, substitutions in positions 101, 120 and 126 were present in the Cases but absent in the Controls (Table 5). Interestingly, in both groups of patients aa substitutions conferring resistance to DAAs were identified, Y93H in two Cases and in 1 Control, L31M in 1 Control (Table 5).
Discussion
In the present study, analyzing the HCV sequences in core, NS3 and NS5A regions in serum, non-cancer liver tissue and in cancer liver tissue of 17 patients with HCC and in the liver tissue of 13 patients without, we tested the variability of HCV genome as the cause of hepatocarcinogenesis.
HCV as a cause of chronic hepatitis and cirrhosis is involved in the promotion phase of hepatocarcinogenesis, indirectly stimulating hepatocellular proliferation.18 However, some studies suggest direct participation of HCV in the oncogenetic process through its structural and non-structural proteins.19 The core protein, the main component of the nucleocapsid, appears to be the main cause of a possible virus-induced proliferation. In fact, it binds to numerous transcription factors, regulating the expression of different genes of the host cell.20 Furthermore, it promotes cell growth and survival by activating the MAPK (Mitogen-Activated Protein Kinase) signalling cascade, by activating proteins such as MEK1, ERK1/2, JNK, MAP kinase p38, and MAP kinase MKP 1.21,22 Moreover, the core protein in the nucleus increases cell proliferation by inhibiting the synthesis of p53, the p21 inhibitor of CDK, and the phosphorylation of pRb.23
In the present study, the phylogenetic analysis showed no differences between the populations of the virus present in the three compartments, suggesting no genetic variability between HCV isolated from cancer tissue and that from non-cancer liver tissue and serum.
Moreover, by the analysis of the mutation patterns of the NS3, NS5A and core regions between patients with or without HCC, a higher aa variability in the NS3 region was observed in the Cases. In addition, we observed that in NS3 and NS5A regions there were some mutations that were repeated more frequently in the patients with HCC than in the Controls, such as in positions 32, 44, 79, 103, 121 and 122 in NS3 and in positions 37 and 54 in the NS5A region. These data may suggest a higher aa variability in patients with HCC than in those without, especially in NS3.
This greater genetic variability found in the NS3 and NS5A regions of the HCV genome of subjects with HCC in our opinion could have a link with the development of liver cancer, since literature studies have shown that the NS3 and NS5A regions are involved in hepatocarcinogenesis. In fact, the NS3 and NS5A proteins may promote hepatocarcinogenesis by its binding to some cellular proteins, such as p21 and p5324,25 and the NS5A protein modulates genes that regulate the cell cycle, promote cell growth and induce aberrant mitosis and chromosomal instability leading to HCC.26–29
However, the patients with HCC were older than those without; thus, these data may be due to a longer infection time that caused a higher aa variability.
There are few studies that analyze the genetic and aa variability of HCV in the cancerous and non-cancerous tissue of HCC patients, with equivocal results.30–34 In the study by Perez et al,31 core and E2 regions were amplified, cloned and sequenced in four patients with HCC who underwent liver transplantation, showing in all the patients a compartmentalization in the three compartments analyzed (tumoral, non-tumoral liver tissue and serum) for both viral regions tested. Harouaka et al32 evaluating 12 patients with HCV-associated HCC (n° = 8) or non-HCC cirrhosis (n° = 4) showed that HCV genetic diversity was significantly higher in livers containing HCC compared with the control non-HCC cirrhotic livers.
Interesting are the results of Sorbo et al35 who investigated the prevalence of resistance-associated aa substitutions (RASs) in plasma, liver tumoral and non-tumoral tissue samples in 18 patients undergoing liver transplant or hepatic resection due to HCC and/or cirrhosis: 6 of 18 patients had at least one RAS in at least one of the compartments analyzed, and nucleotide variability assessed by genetic distance analysis showed higher median values for NS3 and NS5A sequences compared to NS5B in the tissue compartments.
The limit of the present study is the small sample size of the groups. However, we underline that the analysis done, sequencing of NS3, NS5A and core regions both in liver cancer and in non-cancer tissue and in serum of 17 patients with HCC, such as in liver tissue in 13 patients without HCC, although they did not allow to draw unequivocal conclusions, seem to be adequate to a preliminary evaluation of this topic.
In conclusion, from what has been said so far it would seem that what the previous studies have in common with ours is the identification of a greater genetic variability of HCV in tumor tissue compared to non-tumor tissue. Unfortunately, what unites them is also the small size of the sample, so further studies on a wide cohort of patients may better clarify this topic and identify other amino acid substitutions possibly associated with the development of HCC.
Data Sharing Statement
All the data on the study may be requested to the authors, Mariantonietta Pisaturo and Nicola Coppola.
Consent to Participate
All patients signed their informed consent for liver biopsy, the collection and storage of biological samples and for the anonymous use of their data for research purposes.
Consent to Publish
The manuscript has been read and approved by all authors.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare that we have no conflicts of interest related to this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Sagnelli E, Macera M, Russo A, Coppola N, Sagnelli C. Epidemiological and etiological variations in hepatocellular carcinoma. Infection. 2020;48(1):7–17. PMID: 31347138. doi:10.1007/s15010-019-01345-y
3. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi:10.1016/S0140-6736(11)61347-0
4. Gower E, Estes C, Blach S, Razavi‐Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61(1 Suppl):
5. Dhanasekaran R, Bandoh S, Roberts LR. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Res. 2016;5:F1000 Faculty Rev–879. doi:10.12688/f1000research.6946.1
6. Lin MV, King LY, Chung RT. Hepatitis C virus-associated cancer. Annu Rev Pathol. 2014;10:345–370. doi:10.1146/annurev-pathol-012414-040323
7. Banerjee A, Ray RB, Ray R. Oncogenic potential of hepatitis C virus proteins. Viruses. 2010;2:2108–2133. doi:10.3390/v2092108
8. De Giorgi V, Monaco A, Worchech A, et al. Gene profiling, biomarkers and pathways characterizing HCV-related hepatocellular carcinoma. J Transl Med. 2009;7:85. doi:10.1186/1479-5876-7-85
9. Bartsch H, Nair J. Oxidative stress and lipid peroxidation-derived DNA-lesions in inflammation driven carcinogenesis. Cancer Detect Prev. 2004;28:385–391. doi:10.1016/j.cdp.2004.07.004
10. Ogata F, Akuta N, Kobayashi M, et al. Amino acid substitutions in the hepatitis C virus core region predict hepatocarcinogenesis following eradication of HCV RNA by all-oral direct-acting antiviral regimens. J Med Virol. 2018;90(6):1087–1093. doi:10.1002/jmv.25047
11. Nakamoto S, Imazeki F, Fukai K, et al. Association between mutations in the core region of hepatitis C virus genotype 1 and hepatocellular carcinoma development. J Hepatol. 2010;52:72–78. doi:10.1016/j.jhep.2009.10.001
12. Coppola N, Onorato L, Iodice V, et al. Occult HBV infection in HCC and cirrhotic tissue of HBsAg-negative patients: a virological and clinical study. Oncotarget. 2016;7(38):62706–62714. PMID: 27486882; PMCID: PMC5308760. doi:10.18632/oncotarget.10909
13. Coppola N, Potenza N, Pisaturo M, et al. Liver microRNA hsa-miR-125a-5p in HBV chronic infection: correlation with HBV replication and disease progression. PLoS One. 2013;8(7):e65336. PMID: 23843939; PMCID: PMC3701058. doi:10.1371/journal.pone.0065336
14. European Association for the Study of the Liver; European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–943. doi:10.1016/j.jhep.2011.12.001
15. Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19(3):329–338. PMID: 10518312. doi:10.1055/s-2007-1007122
16. Coppola N, Pisapia R, Tonziello G, et al. Improvement in the aetiological diagnosis of acute hepatitis C: a diagnostic protocol based on the anti-HCV-IgM titre and IgG Avidity Index. J Clin Virol. 2009;46(3):222–229. PMID: 19758839. doi:10.1016/j.jcv.2009.08.009
17. Pisaturo M, Starace M, Minichini C, et al. Patients with HCV genotype-1 who have failed a direct-acting antiviral regimen: virological characteristics and efficacy of retreatment. Antivir Ther. 2019;24(7):485–493. PMID: 30758299. doi:10.3851/IMP3296
18. Selimovic D, El-Khattouti A, Ghozlan H, Haikel Y, Abdelkader O, Hassan M. Hepatitis C virus-related hepatocellular carcinoma: an insight into molecular mechanisms and therapeutic strategies. World J Hepatol. 2012;4:342–355. doi:10.4254/wjh.v4.i12.342
19. Kasprzak A, Adamek A. Role of hepatitis C virus proteins (C, NS3, NS5A) in hepatic oncogenesis. Hepat Res. 2008;38:1–26. doi:10.1111/j.1872-034X.2007.00261.x
20. Li HC, Ma HC, Yang CH, Lo SY. Production and pathogenicity of hepatitis C virus core gene products. World J Gastroenterol. 2014;20:7104–7122. doi:10.3748/wjg.v20.i23.7104
21. Tsuchihara K, Hijikata M, Fukuda K, Kuroki T, Yamamoto N, Shimotohno K. Hepatitis C virus core protein regulates cell growth and signal transduction pathway transmitting growth stimuli. Virology. 1999;258:100–107. doi:10.1006/.viro.1999.9694
22. Erhardt A, Hassan M, Heintges T, Haussinger D. Hepatitis C virus core protein induces cell proliferation and activates ERK, JNK, and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology. 2002;292:272–284. doi:10.1006/viro.2001.1227
23. Hassan M, Ghozlan H, bdel-Kader O. Activation of RB/ E2F signaling pathway is required for the modulation of hepatitis C virus core protein-induced cell growth in liver and non-liver cells. Cell Signal. 2004;16:1375–1385. doi:10.1016/j.cellsig.2004.04.005
24. Kwun HJ, Jung EY, Ahn JY, Lee MN, Jang KL. p53- dependent transcriptional repression of p21(waf1) by hepatitis C virus NS3. J Gen Virol. 2001;82:2235–2241. doi:10.1099/0022-1317-82-9-2235
25. Irshad M, Gupta P, Irshad K. Molecular basis of hepatocellular carcinoma induced by hepatitis C virus infection. World J Hepatol. 2017;9(36):1305–1314. PMID: 29359013; PMCID: PMC5756719. doi:10.4254/wjh.v9.i36.1305
26. Ghosh AK, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol. 1999;80:1179–1183. doi:10.1099/0022-1317-80-5-1179
27. Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol. 2001;75:1401–1407. doi:10.1128/JVI.75.3.1401-1407.2001
28. Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kB. Proc Natl Acad Sci U S A. 2001;98:9599–9604. doi:10.1073/pnas.171311298
29. Wu SC, Chang SC, Wu HY, Liao PJ, Chang MF. Hepatitis C virus NS5A protein down-regulates the expression of spindle gene Aspm through PKR-p38 signaling pathway. J Biol Chem. 2008;283(43):29396–29404. PMID: 18728014; PMCID: PMC2662026. doi:10.1074/jbc.M802821200
30. Hu Z, Muroyama R, Kowatari N, Chang J, Omata M, Kato N. Characteristic mutations in hepatitis C virus core gene related to the occurrence of hepatocellular carcinoma. Cancer Sci. 2009;100(12):2465–2468. doi:10.1111/j.1349-7006.2009.01338.x
31. Pérez PS, Di Lello FA, Mullen EG, et al. Compartmentalization of hepatitis C virus variants in patients with hepatocellular carcinoma. Mol Carcinog. 2017;56(2):371–380. PMID: 27163636. doi:10.1002/mc.22500
32. Harouaka D, Engle RE, Wollenberg K, et al. Diminished viral replication and compartmentalization of hepatitis C virus in hepatocellular carcinoma tissue. Proc Natl Acad Sci U S A. 2016;113(5):1375–1380. PMID: 26787866; PMCID: PMC4747736. doi:10.1073/pnas.1516879113
33. El-Shamy A, Pendleton M, Eng FJ, Doyle EH, Bashir A, Branch AD. Impact of HCV core gene quasispecies on hepatocellular carcinoma risk among HALT-C trial patients. Sci Rep. 2016;1(6):27025. doi:10.1038/srep27025
34. Moreira JP, Malta FM, Diniz MA, et al. Interferon lambda and hepatitis C virus core protein polymorphisms associated with liver cancer. Virology. 2016;493:136–141. doi:10.1016/j.virol.2016.03.008
35. Sorbo MC, Carioti L, Bellocchi MC, et al. HCV resistance compartmentalization within tumoral and non-tumoral liver in transplanted patients with hepatocellular carcinoma. Liver Int. 2019;39(10):1986–1998. PMID: 31172639. doi:10.1111/liv.14168
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.