Back to Journals » Infection and Drug Resistance » Volume 12
Genetic characterization of a novel sequence type of multidrug-resistant Citrobacter freundii strain recovered from wastewater treatment plant
Authors Jiang X, Cui X, Liu W , Xu H, Zheng B
Received 26 April 2019
Accepted for publication 16 August 2019
Published 6 September 2019 Volume 2019:12 Pages 2775—2779
DOI https://doi.org/10.2147/IDR.S213525
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xiawei Jiang1, Xinjie Cui1, Wenhong Liu1, Hao Xu2, Beiwen Zheng2
1College of Basic Medical Sciences, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, People’s Republic of China; 2Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, People’s Republic of China
Correspondence: Beiwen Zheng
Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, No. 79 Qingchun Road, Hangzhou 310000, People’s Republic of China
Tel +86 5 718 723 6423
Fax +86 5 718 723 6423
Email [email protected]
Abstract: A multidrug-resistant Citrobacter freundii strain R17 was isolated from a wastewater treatment plant in China. Whole-genome sequencing of strain R17 revealed a new sequence type (ST412) chromosome (length 5,124,258 bp) and an Inc FII (Yp) group plasmid pCFR17_1 (length 206,820 bp). A total of 13 antibiotic-resistance genes (ARGs) that confer resistance to eight different antibiotic groups were encoded by strain R17 and 12 of them were carried by plasmid pCFR17_1. These data and analysis suggest that the environment-derived C. freundii strains may serve as potential sources of ARGs and highlight the need of further surveillance of this bacteria in the future.
Keywords: Citrobacter freundii, multidrug resistant, antibiotic-resistance genes, whole-genome sequencing, wastewater treatment plant, insertion elements
Introduction
Citrobacter freundii is a Gram-negative and facultative anaerobic bacterium which belongs to family Enterobacteriaceae. After its first isolation from soil, C. freundii has been found in various natural habitats, as well as intestines of animals and humans.1–4 Some environment-derived C. freundii strains have potential use in many fields. For example, C. freundii strain JPG1, which was isolated from a gold mining tailing in China, was resistant to heavy metals and capable of removing copper. Thus, it was suggested to have a great potential in the treatment of copper-rich industrial wastewater.5 C. freundii strain IFO 13545 has the ability to produce bioflocculant which can be used in fields of water supply, wastewater treatment, and food production.6 Although C. freundii was previously recognized as a bacterium of low virulence, it has been demonstrated to be the causative agent of a wide spectrum of infections including diarrhea, pneumonia and septicemia.7,8 Moreover, there has been a growing body of literature that reported the plasmid-mediated multidrug resistance in C. freundii, indicating its threat to human health.9,10
Wastewater treatment plants (WWTPs) have been regarded as the major source of antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs).11,12 Previous studies have reported the isolation of several C. freundii strains from WWTPs.13,14 In our investigation of the ARB and ARGs in WWTPs, we obtained a multidrug-resistant strain of C. freundii. This strain (R17) was isolated from the sludge of a WWTP that treats wastewater of a pharmaceutical industry in Taizhou, China. Sludge samples were collected from this WWTP in April 2017. Samples were first cultured overnight in Mueller-Hinton broth without antimicrobial agents, and then the enriched samples were spread on chromID® ESBL agar plates (bioMérieux, Marcy-l’Étoile, France). After incubating at 37°C for 48 hrs, colonies were picked up for identification using both Microflex MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) and 16S rRNA sequencing technology as described previously.4 In the present study, we reported the whole-genome sequencing of C. freundii strain R17. Further investigations were conducted on the genetic features and ARGs associated with its resistance phenotypes.
Antimicrobial susceptibility testing of strain R17 was performed using VITEK 2 system employing panel AST-GN-16 (bioMérieux) with Escherichia coli ATCC 25922 as control. The results were interpreted according to the standards of the Clinical and Laboratory Standards Institute.15 Minimum inhibitory concentration (MIC) of strain R17 indicated its resistance to various types of antibiotics, including ampicillin (MIC ≥32 μg/mL), amoxicillin (MIC ≥32 μg/mL), cefazolin (MIC ≥64 μg/mL), cefoxitin (MIC ≥64 μg/mL), gentamicin (MIC ≥16 μg/mL), ciprofloxacin (MIC≥4 μg/mL), levofloxacin (MIC ≥8 μg/mL) and trimethoprim (MIC ≥320 μg/mL).
Genomic DNA of strain R17 was extracted using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Whole-genome sequencing was performed using Pacific Biosciences RSII platform (PacBio, Menlo Park, CA, USA) and Illumina Hiseq 2000 sequencer (Illumina, San Diego, CA, USA) as described before.16 A chromosome of 5,124,258 bp in length with a GC content of 51.5% and a plasmid (pCFR17_1) of 206,820 bp in length with a GC content of 53.8% were assembled from the sequencing reads using SPAdes software (https://github.com/ablab/spades#sec5).17,18
Annotation with RAST server19 revealed that the chromosome of strain R17 contained 4, 960 protein-coding genes, 42 repeat regions, 85 tRNAs and 25 rRNAs (eight complete 5S-23S-16S rRNA operons and one additional 5S rRNA). Two intact phage regions (one was 48 kb in length and another one was 44.8 kb in length) were predicted from the chromosome using PHASTER.20 One AmpC-type beta-lactamase encoding gene, blaCMY-85, was found in the chromosome of strain R17 using ResFinder 3.121 (Table 1). AmpC enzymes were reported to be involved in expanded-spectrum cephalosporins resistance and were found in the chromosomes of several members of Enterobacterales including C. freundii.22 Up to date, 76.62% (272 of 355) C. freundii strains in NCBI Pathogen Detection database (https://www.ncbi.nlm.nih.gov/pathogens/isolates#/search/) carried blaCMY genes, indicating that blaCMY genes were ubiquitous in C. freundii. To reveal the phylogenetic relationship between strain R17 and other C. freundii strains with published complete chromosome sequences, Average Nucleotide Identities (ANIs) were calculated based on MUMmer using JSpeciesWS.23 Though strain R17 was environmental-derived, it showed high ANI values with the clinical or host-associated strains (Table S1), indicating its potential of being a pathogen. Notably, by searching against C. freundii locus/sequence definitions database (https://pubmlst.org/bigsdb?db=pubmlst_cfreundii_seqdef), we found that strain R17 could not be classified to any known STs, so it was assigned to a new ST, ST412.
 |
Table 1 Distribution of ARGs in C. freundii strain R17 |
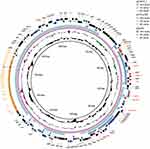
The plasmid typing using PlasmidFinder24 indicated that pCFR17_1 was a Inc FII (Yp) type plasmid. A total of 12 ARGs were predicted in pCFR17_1 using ResFinder 3.121 (Table 1) and these genes were associated with resistance phenotypes of strain R17. By Blastn against NCBI database, we found that pCFR17_1 shared the highest similarity (99.99% identity and 73% coverage) with pCFR-e790 (GenBank accession number CP026241). Plasmid pCFR-e790 was identified in C. freundii strain CFNIH9 which was collected from wastewater of hospital internal pipes in USA.25 These two plasmids shared the core backbone gene loci (Figure 1). Lineal alignment between the two plasmids revealed three highly conserved regions that carried different resistance genes (Figure 2). Mobile elements set adjacent with these genes, indicating the transfer potential of them. One region in pCFR17_1 that contained sul2, catA2 and tet(D) was reversed in pCFR-e790 with two insertion elements (IS110 and IS6) located on both ends of this region, indicating there may be a re-organization mediated by these insertion elements. Sequences encoding for of several mercuric resistance genes were found to lie downstream of blaTEM-1B in both plasmids. This conserved sequence showed high similarity to TnAs3 which belonged to Tn3 composite transposon family. However, a ~24 kb region in pCFR17_1 was absent in pCFR-e790. And, this region exhibited a high similarity to pQnrB4 (GenBank accession number CP028537), a plasmid of Enterobacter hormaechei strain SCEH020042 which was a human-related strain isolated from Sichuan, China (Figure 1). Two insertion elements, IS6 and IS110, located at each ends of this multidrug-resistant region. Between these two insertion elements, there were five ARGs, dfrA2, qnrB4, blaDHA-1 and two sul1 genes. In addition, an operon of phage shock proteins was also carried by this multidrug-resistant region between qnrB4 and blaDHA-1 (Figure 2). Thus, our study is the first isolation of a novel multidrug-resistant environment-derived plasmid pCFR17_1 which showed similarity to both environment-derived plasmid pCFR-e790 and clinical-related plasmid pQnrB4. The emergence of this multidrug-resistant plasmid in strain R17 highlights the potential role of C. freundii as a vector for the dissemination of ARGs in aquatic environments.
 |
Figure 2 Lineal comparison of multidrug-resistant regions of pCFR17_1, pCFR-e790 and pQnrB4. Genes were portrayed by arrows and colored according to their functions. |
In this study, we presented the sequences of the chromosome and the plasmid (pCFR17_1) of a multidrug-resistant C. freundii strain (R17). This is the first report of a novel ST (ST 412) C. freundii strain. The results of this work improved our understanding of the genetic context of C. freundii strains, as well as their antibiotic-resistance phenotypes and genotypes. Environment has been regarded as a potential reservoir of ARB with plasmids conferring resistance. The presence of ARGs, as well as mobile elements in strain R17, suggest that C. freundii may serve as potential source of resistance determinants for more virulent organisms. Thus, C. freundii and their plasmids should be monitored in the future.
Nucleotide sequence accession numbers: The nucleotide sequences of the chromosome and the plasmid (pCFR17_1) of C. freundii strain R17 have been deposited into DDBJ/EMBL/GenBank under accession number CP035276 and CP035277, respectively.
Acknowledgments
We would like to thank Dr. Wenwu Zhang for his assistance in sampling. This work was supported by research grants from the National Key Research and Development Program of China (No. 2016YFD0501105); National Natural Science Foundation of China (Nos. 41406140 and 81741098); and Zhejiang Provincial Natural Science Foundation of China (No. LY17H190003).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Braak H. Onderzoekingen over Vergisting van Glycerine (thesis). Delft, Netherlands: WD Meinema-Utitgeuer; 1928:166.
2. Li M, Li F, Mi Z, et al. Comparative genomics analysis of pTEM-2262, an MDR plasmid from Citrobacter freundii, harboring two unclassified replicons. Future Microbiol. 2018;13(15):1657–1668. doi:10.2217/fmb-2018-0243
3. Osieka V, Grobbel M, Schmoger S, et al. Complete draft genome sequence of an extended-spectrum β-lactamase-producing Citrobacter freundii strain recovered from the intestine of a house sparrow (Passer domesticus) in Germany, 2017. Genome Announc. 2018;6(26):e00599–e00518. doi:10.1128/genomeA.00599-18
4. Xu H, Wang X, Yu X, et al. First detection and genomics analysis of KPC-2-producing Citrobacter isolates from river sediments. Environ Pollut. 2018;235:931–937. doi:10.1016/j.envpol.2017.12.084
5. Wang X, Huang N, Shao J, Hu M, Zhao Y, Huo M. Coupling heavy metal resistance and oxygen flexibility for bioremoval of copper ions by newly isolated Citrobacter freundii JPG1. J Environ Manage. 2018;226:194–200. doi:10.1016/j.jenvman.2018.08.042
6. Baranwal P, Kimura K, Mayilraj S, Negoro S, Takeo M. Draft genome sequence of a bioflocculant-producing bacterium, Citrobacter freundii IFO 13545. Microbiol Resour Announcements. 2019;8(29):e00524–e00519. doi:10.1128/MRA.00524-19
7. Lan R, Xu J. Genetic diversity, multidrug resistance and virulence of Citrobacter freundii from diarrheal patients and healthy individuals. Front Cell Infect Microbiol. 2018;8:233. doi:10.3389/fcimb.2018.00026
8. Anderson MT, Mitchell LA, Zhao L, Mobley HL. Citrobacter freundii fitness during bloodstream infection. Sci Rep. 2018;8(1):11792. doi:10.1038/s41598-018-30196-0
9. Ouyang J, Sun F, Zhou D, et al. Comparative genomics of five different resistance plasmids coexisting in a clinical multi-drug resistant Citrobacter freundii isolate. Infect Drug Resist. 2018;11:1447. doi:10.2147/IDR.S165818
10. Wu W, Espedido B, Feng Y, Zong Z. Citrobacter freundii carrying bla KPC-2 and bla NDM-1: characterization by whole genome sequencing. Sci Rep. 2016;6:30670. doi:10.1038/srep30670
11. Obayiuwana A, Ogunjobi A, Yang M, Ibekwe M. Characterization of bacterial communities and their antibiotic resistance profiles in wastewaters obtained from pharmaceutical facilities in Lagos and Ogun States, Nigeria. Int J Environ Res Public Health. 2018;15:7. doi:10.3390/ijerph15061188
12. Zhai W, Yang F, Mao D, Luo Y. Fate and removal of various antibiotic resistance genes in typical pharmaceutical wastewater treatment systems. Environ Sci Pollut Res. 2016;23(12):12030–12038. doi:10.1007/s11356-016-6350-9
13. Duceppe M-O, Phipps-Todd B, Carrillo C, Huang H. Draft genome sequences of eight Canadian Citrobacter braakii and Citrobacter freundii strains. Microbiol Resour Announcements. 2019;8(27):e00273–e00219. doi:10.1128/MRA.00273-19
14. Aristizábal-Hoyos AM, Rodríguez EA, Arias L, Jiménez JN. High clonal diversity of multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in a wastewater treatment plant. J Environ Manage. 2019;245:37–47. doi:10.1016/j.jenvman.2019.05.073
15. Performance CLSI. Standards for Antimicrobial Susceptibility Testing.
16. Jiang X, Liu W, Zheng B. Complete genome sequencing of Comamonas kerstersii 8943, a causative agent for peritonitis. Sci Data. 2018;5:180222. doi:10.1038/sdata.2018.187
17. Nurk S, Bankevich A, Antipov D, et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F, Zhang X (editors). Research in Computational Molecular Biology. RECOMB 2013. Lecture Notes in Computer Science, vol 7821. Heidelberg: Springer, Berlin; 2013.
18. Antipov D, Korobeynikov A, McLean JS, Pevzner PA. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 2015;32(7):1009–1015. doi:10.1093/bioinformatics/btv688
19. Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi:10.1186/1471-2164-9-75
20. Arndt D, Marcu A, Liang Y, Wishart DS. PHAST, PHASTER and PHASTEST: tools for finding prophage in bacterial genomes. Brief Bioinform. 2017. doi:10.1093/bib/bbx121
21. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
22. Meini S, Tascini C, Cei M, Sozio E, Rossolini GM. AmpC β-lactamase-producing Enterobacterales: what a clinician should know. Infection. 2019;47(3):363-375.
23. Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2015;32(6):929–931. doi:10.1093/bioinformatics/btv681
24. Carattoli A, Zankari E, Garcìa-Fernandez A, et al. PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother. 2014;58(7):3895-3903.
25. Weingarten RA, Johnson RC, Conlan S, et al. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. MBio. 2018;9(1):e02011–e02017. doi:10.1128/mBio.02011-17
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.