Back to Journals » Infection and Drug Resistance » Volume 15
Genetic and Phenotypic Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Isolates from a Tertiary Hospital in Beijing
Authors Ni Q, Yao X, Li J, Ma J, Wang K, Liu X , Li P, Yang L, Li P, Li S
Received 2 November 2022
Accepted for publication 9 December 2022
Published 19 December 2022 Volume 2022:15 Pages 7503—7508
DOI https://doi.org/10.2147/IDR.S395920
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Qin Ni,1,2,* Xingwei Yao,3,* Jinhui Li,2,* Jinghan Ma,3,* Kaiying Wang,2 Xiong Liu,2 Peihan Li,2 Lang Yang,2 Peng Li,2 Shenlong Li1,2
1Epidemiology, Zhengzhou University, Zhengzhou, People’s Republic of China; 2Biosafety, Chinese PLA Center for Disease Control and Prevention, Beijing, People’s Republic of China; 3Medical Clinical Laboratory, Dong Zhimen Hospital Beijing University of Chinese Medicine, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Peng Li; Shenlong Li, Email [email protected]; [email protected]
Objective: Klebsiella pneumoniae is a common multidrug-resistant pathogen that jeopardizes the health of hospitalized patients. We aimed to study the phenotypic and genotypic characteristics of carbapenem-resistant K. pneumoniae (CRKP) isolates from a hospital in Beijing.
Methods: Twenty-four CRKP clinical isolates were collected within a half-year to investigate antimicrobial resistance and genomic characteristics. Illumina and Nanopore sequencing were performed to assemble and annotate genomes.
Results: All strains were multi-drug resistant. Twenty-two strains carried the blaKPC-2 gene and two harbored blaNDM-5. Multilocus sequence type(MLST) analysis identified five sequence types; most isolates belonged to ST11. Three strains were isolated from the same patient; each carried a different plasmid replicon, either IncFII (pHN7A8), IncX, or IncFIB (K).
Conclusion: This study furthers the understanding of CRKP antimicrobial resistance genotypes, and may facilitate the control of nosocomial infections caused by antimicrobial-resistant pathogens.
Keywords: Klebsiella pneumoniae, drug resistance, carbapenemase, ST11, blaKPC-2
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) comprise a set of Gram-negative bacteria that cause serious nosocomial infections, including pneumonia, meningitis, liver abscess, wound and urinary tract infections, especially in immunodeficient patients.1,2 The rising prevalence and increasing multidrug resistance of CRKP are of critical concern.3 A World Health Organization report on antimicrobial resistance disclosed that CRKP has a worldwide geographic distribution, and that the prevalence of CRKP among K. pneumoniae isolates exceeds 50% in some patient groups.4 For example, carbapenem resistance among K. pneumoniae isolates in Greece increased from 0% in 2003 to 38.3% in 2010.5 Surveillance reports from CHINET show that the prevalence of meropenem resistance among K. pneumoniae isolates in China has increased annually, from 2.9% in 2005 to 26.3% in 2018.6
Carbapenems are used to treat severe infections caused by multidrug-resistant Enterobacteriaceae (including K. pneumoniae). The emergence of carbapenemases and extended-spectrum beta-lactamases (ESBL) in K. pneumoniae has reduced bacterial sensitivity to almost all beta-lactams,7 which confounds treatment and jeopardizes clinical outcomes.
blaKPC-2 is the major carbapenemase gene in Enterobacteriaceae isolated from Chinese adults and children.6,8 blaNDM-positiveK. pneumoniae isolates often produce metallo-β-lactamase9 and demonstrate high-level resistance to all carbapenems.10 Understanding the types and distribution of K. pneumoniae antibiotic resistance genes (ARGs) facilitates hospital infection control and deserves attention.11
We employed Illumina and Nanopore sequencing to analyze the genomic epidemiology of CPKP clinical isolates. Our findings characterized the genomic and resistance features of K. pneumoniae and revealed an evolving genotype in isolates from a single patient, and may guide prevention and treatment.
Methods
Bacterial Classification and Antimicrobial Susceptibility Testing
Samples were collected in a tertiary hospital in Beijing, China, from June to December 2021. Twenty-four CRKP isolates were obtained from sputum (11/24), urine (7/24), blood (3/24), catheters (1/24), secretions (1/24) and throat swabs (1/24). The samples were collected from 15 (62.5%) men and 9(37.5%) women between 50 and 94 years old. A fully automated analysis system (PHOENIX-100 [BD, USA]) was used for species identification and antimicrobial susceptibility testing. Susceptibilities to 18 commonly used antibacterial agents were determined; these included ampicillin, amoxicillin/clavulanate, ampicillin/sulbactam, piperacillin/tazobactam, cefazolin, cefepime, cefotaxime, ceftazidime, meropenem, imipenem, gentamicin, amikacin, ciprofloxacin, moxifloxacin, levofloxacin, trimethoprim/sulfamethoxazole, tetracycline, and aztreonam. Results were interpreted according to the breakpoints and reporting method of the Clinical Laboratory Standards Institute (2021 version, https://clsi.org).
Whole-Genome Sequencing Using Illumina and Nanopore
Genomic DNA was extracted from cultured bacteria. The Illumina sequencing library was prepared using NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England BioLabs, CA, USA) according to the manufacturer’s instructions. Sequencing was performed on the Illumina NovaSeq 6000 platform at Novogene company with a strategy of pair-end 150bp. Nanopore sequencing libraries were prepared using the Ligation Sequencing Kit SQK-LSK109 (Oxford Nanopore Technologies, Cambridge, UK). The prepared libraries were sequenced on a MinION MK1B sequencer (Oxford Nanopore Technologies, Cambridge, UK) with an R9.4 flow cell.
Dry Lab: Bioinformatics Analysis
The term dry lab denotes the bioinformatic portion of the technology.12 Illumina and Nanopore sequencing data were filtered by Fastp13 (version 0.19.7) and NanoFilt (v2.8.0). Hybrid assembly of clean data was conducted with Unicycler (v0.4.8).14 The ARGs of CRKP were identified using the ABRicate (1.0.1) and Resfinder15 databases (https://cge.food.dtu.dk/services/ResFinder/). Kleborate (v2.0.1)16 was used for multilocus sequence typing (MLST). Plasmids were identified using the PlasmidFinder17 database. Genomes were annotated using Prokka (1.14.6).18 Core genome alignments were created by Roary,19 and maximum likelihood phylogenetic trees were generated using RAxML-NG20 (v1.0.3), visualized using FigTree (v1.4.4) and the R ggplot2 package. Plasmid genome circle map visualization using proksee (https://proksee.ca).
Results
CRKP Isolation and Antimicrobial Susceptibility Testing
From June to December 2021, 24 CRKP isolates were obtained from sputum (11/24), urine (7/24), blood (3/24), catheters (1/24), secretions (1/24), and throat swabs (1/24). Specimens were primarily from the Intensive Care Unit (ICU) (17/24), followed by neurology (3/24), neonatology (2/24) and nephrology (2/24) wards. All strains were ESBL-positive. Three strains (BJ10-0010, BJ10-0013 and BJ10-0015) were isolated from urine, blood, and sputum of the same ICU patient in June, September, and October 2021, respectively. The samples were collected from 15 (62.5%) men and 9(37.5%) women between 50 and 94 years old. Most strains were resistant to multiple agents, including meropenem and imipenem (Table 1). All 24 strains (100%) were resistant to ampicillin, cefazolin, amoxicillin/clavulanate, ampicillin/sulbactam, meropenem, piperacillin/tazobactam, cefepime, cefotaxime and ceftazidime, but most (66.67%) were sensitive to amikacin. Interestingly, the three strains from the same patient showed slightly different levels of imipenem resistance. The MIC values of the 22 strains are provided in Supplementary Table S1.
 |
Table 1 Antimicrobial Susceptibility of K. pneumoniae Isolates |
Genomic Characterization of CRKP
The genomes of 24 CRKP strains were obtained by hybrid assembly using Illumina and nanopore sequencing, and have been deposited in NCBI under the BioProject accession number PRJNA874878. Whole genome MLST identified five sequence types. Most strains belonged to ST11 (19/24), and others belonged to ST15 (2/24), ST37 (1/24), ST403 (1/24) and ST1530 (1/24). Genomic annotation identified diverse carbapenemase- and β-lactamase-encoding ARGs. The blaLAP-2 (13/24), blaCTX-M-65 (7/24) and blaSHV-12 (7/24) genes were chromosomal, while blaKPC-2 (13/24), blaTEM-1B (12/24), blaOXA-1 (8/24), blaNDM-5 (2/24) blaSHV-12 (2/24) and blaCTX-M-3 (1/24) blaCTX-M-65 (1/24) were plasmid-encoded. The distribution of ARGs of strains of different STs is shown in Figure 1.
Distribution of Antibiotic Resistance Genes Among Plasmid Replicon Types
Plasmid incompatibility groups were determined according to replication sequences. The strains contained multiple plasmid replicon types, most of which distributed in ST11 strains. All ST11 strains containing plasmid replicons IncFII (pHN7A8). IncFIB (K), IncFIB (pKPHS1) or IncFIB (pNDM-Mar) were distributed exclusively in subclade1, while IncR was carried only in subclades 2 and 3. Twenty-five plasmids contained single replicons, and 14 carried multiple replicons. On average, each single-replicon plasmid carried 1.52 ARGs, and each multi-replicon plasmid encoded 2.71 ARGs. Plasmids containing multiple-replicons IncFIB(K)/IncFII(pHN7A8) and IncFIB(K)/IncFIB(pKPHS1) carried blaKPC-2, blaTEM-1B, and rmtB. The plasmids of BJ10-0013 included the IncFII (K) replicon and encoded eight ARGs, the most of all plasmids.
Comparison of BJ19-0010, BJ19-0013 and BJ19-0015 from a Single Patient
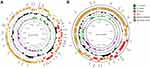
These three strains were isolated from different samples from the same patient. Interestingly, BJ19-0010 and BJ19-0015 belonged to ST11 and were resistant to ciprofloxacin and levofloxacin, while strain BJ19-0013 belonged to ST1503 and was fluoroquinolone-sensitive. In the phylogenetic tree, BJ19-0010 and BJ19-0015 fell in subclade 1, while BJ19-0013 comprised a distinct branch. Strains BJ19-0010, BJ19-0013 and BJ19-0015 had diverse plasmid replicons. Interestingly, BJ19-0010 and BJ19-0015 shared IncFII (pHN7A8), while BJ19-0010 carries IncFIB(K)/IncFIB(pNDM-Mar) and BJ19-0015 carries IncFIB (K). Both BJ19-0010 and BJ19-0015 carried blaKPC-2, blaLAP-2 and blaOXA-1. The blaKPC-2 and blaOXA-1 genes were plasmid-encoded, while the blaLAP-2 gene was located on the chromosome. Strain BJ19-0010 also contained two copies of blaTEM-1B that were located on different plasmids. However, BJ19-0013 carries IncX3 and IncFII(K), which are different from the other two strains. The hybrid assembly showed that the IncFII(K) plasmid pBJ1913-TEM had a length of 127942bp and carries the β- Lactams ARGs blaTEM-1B and blaCTX-M-3(Figure 2A). The other plasmid, pBJ1913-NDM5, had a length of 46068bp and carries the carbapenem-resistance gene blaNDM-5 (Figure 2B), which is nearly identical with plasmid pR15_NDM-5 in Escherichia coli strain R15 isolated from wastewater treatment plant in China.
Discussion
Carbapenems are usually effective for the treatment of drug-resistant bacterial infections. However, CRKP can express plasmid-encoded carbapenemases, which inactivate most β-lactam antibiotics and confer carbapenem resistance.21 Our study showed that 24 CRKP strains were resistant to most tested drugs. The resistance rate to fluoroquinolones was relatively high, which was consistent with the results of a previous study.22 Three strains were isolated from different samples from the same patient within 4 months and carried different carbapenemase genes, which were located on separate plasmids. Genome annotation revealed that Tn3 was located upstream of the blaKPC-2 gene on chromosomes of BJ19-0010 and BJ19-0015, while Tn552 was located downstream of blaNDM-5 in BJ19-0013. The Tn3 transposon promotes the dissemination of AMR,23 and Tn552 has a co-integrate resolution system homologous to Tn3 family elements,24 which may have facilitated plasmid and ARG exchange among these three strains. A previous study showed that blaKPC−2-positive K. pneumoniae isolates in China belonged primarily to ST11.25 In this study, the majority of CRKP strains belonged to ST11, all of which contained blaKPC-2. Most (68.42%, 19/24) ST11 strains were isolated from ICU patients; consequently, the monitoring of AMR strains in ICUs should be strengthened. In K. pneumoniae, the plasmid with an IncR replicon was identified as a multidrug resistance vector with variable ARGs.26 ST11 strains in this study contained the IncR and IncFII (pHN7A8) replicons with blaCTX-M-65, blaTEM-1B, blaKPC-2, and rmtB genes.
Conclusion
Our study revealed the prevalence of CRKP strains isolated primarily from ICU patients. All 24 strains (100%) were resistant to β- Lactams and carbapenems and carried diverse plasmids with different ARGs. The rapid dissemination of ST11 strains highlights the necessity of strengthening CRKP surveillance and may provide a theoretical basis for infection control and treatment.
Data Sharing Statement
The genomes of 24 CRKP strains have been deposited in NCBI with the BioProject accession number PRJNA874878.
Ethics Approval and Informed Consent
The institutional ethics committees of Dong zhimen Hospital Beijing University of Chinese Medicine approved the study. As all data were anonymously collected and interpreted, the institutional ethics committees waived the need for written informed consent from the participants. The study was also supervised by Chinese PLA Center for Disease Control and Prevention.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The work was supported by the National Key Research and Development Program of China (2021YFC2301000) and the National Science and Technology Major Project (2018ZX10201001).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
2. Ahmadi M, Ranjbar R, Behzadi P, et al. Virulence factors, antibiotic resistance patterns, and molecular types of clinical isolates of Klebsiella Pneumoniae. Expert Rev Anti Infect Ther. 2022;20(3):463–472. doi:10.1080/14787210.2022.1990040
3. Lee CR, Lee JH, Park KS, et al. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi:10.3389/fmicb.2016.00895
4. Antimicrobial resistance, global report on surveillance; 2014. Available from: https://www.who.int/publications/i/item/9789241564748.
5. Zagorianou A, Sianou E, Iosifidis E, et al. Microbiological and molecular characteristics of carbapenemase-producing Klebsiella pneumoniae endemic in a tertiary Greek hospital during 2004–2010. Euro Surveill. 2012;17(7):20088.
6. China antimicrobial surveillance network, 2021.Available from: http://www.chinets.com/Data/AntibioticDrugFast.
7. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9(4):228–236. doi:10.1016/S1473-3099(09)70054-4
8. Han R, Shi Q, Wu S, et al. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
9. Behzadi P, García-Perdomo HA, Karpiński TM, et al. Metallo-ß-lactamases: a review. Mol Biol Rep. 2020;47(8):6281–6294. doi:10.1007/s11033-020-05651-9
10. Fu B, Yin D, Sun C, et al. Clonal and horizontal transmission of bla(NDM) among Klebsiella pneumoniae in children’s intensive care units. Microbiol Spectr. 2022;10(4):e0157421. doi:10.1128/spectrum.01574-21
11. Ahmadi Z, Noormohammadi Z, Ranjbar R, et al. Prevalence of tetracycline resistance genes tet (A, B, C, 39) in Klebsiella pneumoniae isolated from Tehran, Iran. Iran J Med Microbiol. 2022;16(2):141–147. doi:10.30699/ijmm.16.2.141
12. Behzadi P, Ranjbar R. DNA microarray technology and bioinformatic web services. Acta Microbiol Immunol Hung. 2019;66(1):19–30. doi:10.1556/030.65.2018.028
13. Chen S, Zhou Y, Chen Y, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi:10.1093/bioinformatics/bty560
14. Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005595
15. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
16. Lam MMC, Wick RR, Watts SC, et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(1):4188. doi:10.1038/s41467-021-24448-3
17. Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi:10.1128/AAC.02412-14
18. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
19. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi:10.1093/bioinformatics/btv421
20. Kozlov AM, Darriba D, Flouri T, et al. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi:10.1093/bioinformatics/btz305
21. Jeon JH, Lee JH, Lee JJ, et al. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int J Mol Sci. 2015;16(12):9654–9692. doi:10.3390/ijms16059654
22. Ahmadi Z, Noormohammadi Z, Behzadi P, et al. Molecular detection of gyrA mutation in clinical strains of Klebsiella pneumoniae. Iran J Public Health. 2022;51(10):2334–2339. doi:10.18502/ijph.v51i10.10992
23. Nicolas E, Lambin M, Dandoy D, et al. The Tn3-family of replicative transposons. Microbiol Spectr. 2015;3(4). doi:10.1128/microbiolspec.MDNA3-0060-2014
24. Rowland SJ, Dyke KG. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4(6):961–975. doi:10.1111/j.1365-2958.1990.tb00669.x
25. Qi Y, Wei Z, Ji S, et al. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–312. doi:10.1093/jac/dkq431
26. Yu X, Zhang W, Zhao Z, et al. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae isolates with focus on antimicrobial resistance. BMC Genom. 2019;20(1):822. doi:10.1186/s12864-019-6225-9
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.