Back to Journals » Cancer Management and Research » Volume 13
GATA1-Activated HNF1A-AS1 Facilitates the Progression of Triple-Negative Breast Cancer via Sponging miR-32-5p to Upregulate RNF38
Received 27 July 2020
Accepted for publication 24 November 2020
Published 11 February 2021 Volume 2021:13 Pages 1357—1369
DOI https://doi.org/10.2147/CMAR.S274204
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Kenan Onel
Jingyu Yang, Heng Niu, Xin Chen
Department of Breast Thyroid Surgery, The First People’s Hospital of Yunnan Province, Kunming, 650032, People’s Republic of China
Correspondence: Jingyu Yang
Department of Breast Thyroid Surgery, The First People’s Hospital of Yunnan Province, No. 157 Jinbi Road, Kunming, 650032, People’s Republic of China
Email [email protected]
Background: Triple-negative breast cancer (TNBC) is a highly invasive subtype of breast cancer with a high mortality rate. Recently, long non-coding RNAs (lncRNAs) are confirmed to modulate the progression of assorted cancers, including TNBC. However, the functions of lncRNA HNF1 homeobox A antisense RNA 1 (HNF1A-AS1) in TNBC are still unclear.
Aim: We aimed to investigate the function and mechanism of HNF1A-AS1 in TNBC.
Methods: The expression of genes in TNBC cells was tested by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot. In vitro loss-of-function assays and in vivo xenograft experiments were conducted for evaluating the impact of HNF1A-AS1 on TNBC progression. RNA pull-down, luciferase reporter and RNA immunoprecipitation (RIP) assays were utilized for assessing the correlations between molecules.
Results: We discovered that HNF1A-AS1 was highly expressed in TNBC tissues and cells. Knockdown of HNF1A-AS1 restrained cell proliferation but accelerated cell apoptosis. Besides, GATA-binding protein 1 (GATA1) activated HNF1A-AS1 transcription in TNBC. MicroRNA-32-5p (miR-32-5p) was slowly expressed in TNBC cells and sponged by HNF1A-AS1, and its overexpression hinders TNBC cell growth. Ring finger protein 38 (RNF38) was verified as the target of miR-32-5p, and HNF1A-AS1 was a competing endogenous RNA (ceRNA) of RNF38 through sponging miR-32-5p. Rescue experiments indicated that upregulation of RNF38 reversed the inhibited impacts of silencing HNF1A-AS1 on TNBC cell growth.
Conclusion: GATA1-activated HNF1A-AS1 facilitated TNBC progression via miR-32-5p/RNF38 axis. The findings may provide new roads for developing targeted therapies of TNBC.
Keywords: HNF1A-AS1, miR-32-5p, RNF38, triple-negative breast cancer
Introduction
As we all know, breast cancer is the most common type of malignant tumor in women, which seriously harms women’s health.1 It has become the major cause of cancer-related deaths among women and TNBC-caused deaths account for 20% of women’s deaths in the whole world.2 Evidences have indicated that breast cancer is a heterogeneous disease and it can be divided into different pathological subtypes.3 Among them, triple-negative breast cancer (TNBC) acts as a subtype, accounting for about 20% of all breast cancer cases.4 A major feature of TNBC is the lack of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2).5 Because of the highly aggressive nature of TNBC, the prognosis and survival of patients are very poor. At present, common treatment methods such as surgery, chemotherapy, and radiotherapy are adopted as treatment strategies for TNBC, but the effects are not satisfactory.6 Hence, investigating the molecular mechanism of TNBC can conduce to discover new therapeutic directions and elevate patient survival.
Long non-coding RNAs (lncRNAs) are defined as RNA transcripts that are more than 200 nucleotides in length and cannot encode proteins.7 Lots of lncRNAs have been indicated to modulate sundry biological behaviors in cancer cells, such as cell proliferation, apoptosis and migration.8 Of note, lncRNAs with abnormal expression are considered as crucial factors in cancer development. For example, SNHG20 was proved to be upregulated in non-small cell lung cancer cells and the inhibition of it could suppress cell proliferation and accelerate cell apoptosis in this cancer by serving as a miR-154 sponge.9 Also, ASAP1-IT1 could enhance cancer stemness and its upregulation could predict bad prognosis in patients with bladder cancer.10 Moreover, AWPPH expedited hepatocellular carcinoma progression via YBX1 and might be served as a prognostic biomarker.11 HNF1 homeobox A antisense RNA 1 (HNF1A-AS1) is a lncRNA that has been confirmed to exert regulatory functions in several cancers. For instance, HNF1A-AS1 was reported to accelerate cell growth in bladder cancer.12 Besides, HNF1A-AS1 could also expedite cell proliferation and invasion by its regulation on miR-17-5p in non-small cell lung cancer.13 Moreover, HNF1A-AS1 was suggested to accelerate carcinogenesis in colorectal cancer by inducing the activation of Wnt/β-catenin signaling pathway.14 Nevertheless, it is not researched in TNBC.
In recent years, increasing researches have indicated that lncRNAs could regulate the progression of cancers through sponging microRNAs (miRNAs) to modulate the expression of messenger RNAs (mRNAs).15 This regulatory mechanism is called competing endogenous RNA (ceRNA) mechanism which has been frequently discovered in cancers.16 For example, HNF1A-AS1 could accelerate non-small cell lung cancer progression by acting as a ceRNA to sponge miR-149-5p and upregulate Cdk6.17 However, whether HNF1A-AS1 can play the role of ceRNA in TNBC remains to be deeply studied.
MiRNAs are a kind of endogenous small noncoding RNA molecules with 19–26 nucleotides in length. As mentioned above, a flow of researches have confirmed that lncRNAs exert their functions by interacting with miRNAs.18 In addition, miRNAs can also play the different roles on cancer process. For example, miR-32-5p was confirmed to modulate the progression of colorectal cancer and cervical cancer.19,20 Meanwhile, mRNAs can be targeted by miRNAs and the encoded proteins become the crucial factors in the regulation of cancers.21 As an example, ring finger protein 38 (RNF38) has been supported as an oncogene in some cancers such as hepatocellular carcinoma, gastric cancer and non-small cell lung cancer.22–24
In the current study, we proposed to search the function and molecular mechanism of HNF1A-AS1 in TNBC. We firstly detected its expression in TNBC tissues and cells. Further, functional assays were designed for evaluating the role of HNF1A-AS1 in TNBC cells. Besides, the interaction among HNF1A-AS1, miR-32-5p and RNF38 was also investigated by mechanistic assays. Our finding may offer a novel idea for the investigation of TNBC treatment.
Materials and Methods
Clinical Specimens
The TNBC tissues and paired para-tumor tissues were surgically acquired from 28 TNBC patients who received no other treatment before operation. All samples were processed with liquid nitrogen immediately after excision and then stored at −80°C until use. Each patient signed the informed consent prior to this study. Tissues were collected and applied under the approval of the Ethics Committee of the First People’s Hospital of Yunnan Province.
Cell Lines
Human TNBC cell lines (MDA-MB-453, MDA-MB-468, MDA-MB-231, BT-20 and MDA-MB-436) and the normal human breast epithelial cell line (MCF-12F), were available from American Type Culture Collection (ATCC; Manassas, VA, USA) and then incubated under 37°C in 5% CO2. Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% penicillin/streptavidin and 10% fetal bovine serum (FBS), was acquired from Gibco (Rockville, MD, USA) for cell culture purposes.
Total RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from cell samples by use of TRIzol Reagent (Invitrogen, Carlsbad CA, USA), and then converted into complementary DNA (cDNA) by use of PrimeScript Reverse Transcriptase Kit (Takara, Shiga, Japan). Gene expression was then quantified by qPCR with SYBR Green PCR Kit (Takara) and calculated based on the 2−ΔΔCt method. U6 snRNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA served as the endogenous control. The experiment was repeated at least three times.
Cell Transfection
The specific short hairpin RNAs (shRNAs) to HNF1A-AS1 and the nonspecific negative control (NC) shRNAs were designed and constructed by Genechem (Shanghai, China). The full-length cDNA sequences of GATA1 and RNF38 were separately inserted into the pcDNA3.1 vectors for gene overexpression, with empty vectors utilized as NC. The miR-32-5p mimics, miR-32-5p inhibitor and their corresponding NCs were acquired from GenePharma (Shanghai, China). MDA-MB-436 and BT-20 cells in 6-well plates (1 × 106 cells/well) were transfected in the presence of Lipofectamine 3000 (Invitrogen) for 48 h, and then harvested for analysis. The experiment was repeated at least three times.
Colony Formation Assay
Transfected MDA-MB-436 and BT-20 cells in 6-well plates were seeded at the cell density of 500/well for 14 days. Then, the resulting colonies were fixed for 30 min in 4% paraformaldehyde (PFA) and stained for 5 min in 0.5% crystal violet solution, finally imaged and counted manually. The experiment was repeated at least three times.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
EdU assay was undertaken in transfected MDA-MB-436 and BT-20 cells using BeyoClick™ EdU Cell Proliferation Kit (Beyotime, Shanghai, China) with Alexa Fluor 594. After nuclear detection in 4ʹ,6-diamidino-2-phenylindole (DAPI) solution, cells were analyzed using the fluorescence microscope (Olympus, Tokyo, Japan). The experiment was repeated at least three times.
JC-1 Assay
Cell apoptosis was examined via the change in mitochondrial membrane potential (Δψm) using JC-1 assay. After transfection, 1 mL of cell suspension in 6-well plates were incubated for 30 min with 2.5 µg/mL of JC-1 dye at 37°C, and then analyzed by the microscope (Olympus). The experiment was repeated at least three times.
Terminal Deoxynucleotidyl Transferase (TdT) dUTP Nick-End Labeling (TUNEL) Assay
TUNEL assay was accomplished in transfected MDA-MB-436 and BT-20 cells with the application of TUNEL reagents (Merck KGaA, Darmstadt, Germany). After washing and fixing, cells were double-stained in TUNEL and DAPI, and then assayed via the fluorescence microscopy (Olympus). The experiment was repeated at least three times.
Flow Cytometry Analysis
The apoptosis of MDA-MB-436 and BT-20 cells were studied using Annexin V-FITC/PI double staining kit (Invitrogen). Cell samples were stained in the dark for 15 min, and then studied by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA). The experiment was repeated at least three times.
In vivo Xenograft Model
To simulate tumor growth in vivo, TNBC cells transfected with sh-NC or sh-HNF1A-AS1#1 were injected into the back of mice (4–6 weeks old; female) that were purchased from SLRC Laboratory Animal Center (Shanghai, China). After that, tumor volume was monitored every 4 days. After tumors grew for 28 days, the mice were sacrificed and the xenografts were isolated, photographed and weighed. The experiment was approved by the Institutional Animal Care and Use Committee of the First People’s Hospital of Yunnan Province.
Luciferase Reporter Assay
The sequences of HNF1A-AS1 promoter region covering wild-type (WT) or mutant (Mut) GATA1 binding sites were cloned into the pGL3-basic vector for promoter analysis, and then the recombinant vectors were co-transfected with pcDNA3.1 or pcDNA3.1/GATA1 into MDA-MB-436 and BT-20 cells for 48 h. The full-length HNF1A-AS1 or RNF38 3ʹUTR (3ʹ-untranslated region) fragments covering wild-type or mutant miR-32-5p binding sites were inserted into the pmirGLO dual-luciferase vectors for interaction analysis. Then, the constructs were co-transfected with miR-32-5p mimics or NC mimics into MDA-MB-436 and BT-20 cells for 48 h. The luciferase activity was studied using the dual-luciferase reporter assay system (Promega, Madison, WI). The experiment was repeated at least three times.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was undertaken in MDA-MB-436 and BT-20 cells by the EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA, USA). The cells were fixed in 4% PFA for 15 min’s crosslink, and then subjected to ultrasonic to shear DNA into 500 bp of fragments. Then, the fragments were subjected to immunoprecipitation for 6 h with GATA1 antibody or control immunoglobulin G (IgG) antibody. The magnetic beads (30 μL) were added for collecting the precipitated chromatin fragments, followed by the fragments analyzed via qRT-PCR. The experiment was repeated at least three times.
Subcellular Fractionation
The subcellular localization of HNF1A-AS1 in MDA-MB-436 and BT-20 cells was determined by subcellular fractionation as per the protocol of PARIS™ Kit (Invitrogen). GAPDH and U6 were separately defined as the endogenous control for cytoplasmic or nuclear RNAs. The experiment was repeated at least three times.
Fluorescence in situ Hybridization (FISH) Assay
The RNA FISH probe designed for HNF1A-AS1 was acquired from Ribobio (Guangzhou, China). MDA-MB-436 and BT-20 cells were fixed in 4% PFA, digested and air-dried, and then subjected to incubation in hybridization buffer with HNF1A-AS1 probes. After Hoechst staining, cells were studied by the fluorescence microscope (Olympus). The experiment was repeated at least three times.
RNA Pull-Down Assay
RNA–RNA interaction was assessed by RNA pull-down by use of Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA) following the user guide. Cell lysates obtained via the cell lysis buffer were mixed overnight with biotinylated HNF1A-AS1 probes or miR-32-5p sequences with wild-type or mutant seed region, or the corresponding control probes. After that, the mixtures were added with magnetic beads, and then the captured RNAs were analyzed by qRT-PCR. The experiment was repeated at least three times.
RNA Immunoprecipitation (RIP)
RIP assay was performed in MDA-MB-436 and BT-20 cells by applying the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit following the standard method (Millipore, Bedford, MA). Firstly, cells were lysed via RIP lysis buffer. Then, cell lysates were cultured overnight in RIP buffer covering magnetic beads and human argonaute RISC catalytic component 2 (Ago2) antibody or control IgG antibody. Finally, the precipitated RNAs were studied using qRT-PCR. The experiment was repeated at least three times.
Western Blot
Cell samples were lysed in radio-immunoprecipitation assay (RIPA) lysis buffer. Then, the collected proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then shifted onto polyvinylidene fluoride (PVDF) membranes. After sealing by 5% nonfat milk, PVDF membranes were probed with the specific primary antibodies at 4°C all night, and then with the secondary antibodies at room temperature for 2 h after washing in Tris-buffered saline Tween-20 (TBST). All of the used antibodies were procured from Abcam (Cambridge, MA). The protein signal was detected via the enhanced chemiluminescence (ECL) luminous liquid (Pierce, Rockford, IL, USA). The experiment was repeated at least three times.
Statistical Analysis
Data were exhibited as the means ± standard deviation (SD) of three individually implemented assays. Statistical analyses of this study were undertaken with Student’s t-test or one-way analysis of variance (ANOVA) by use of GraphPad Prism 7 (La Jolla, CA), with p-values below 0.05 as the cutoff value.
Results
HNF1A-AS1 Was Upregulated in TNBC and Silencing HNF1A-AS1 Restrained the Tumorigenesis of TNBC
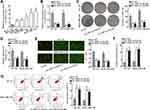
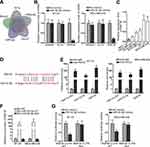
In lots of researches, the imbalance of lncRNA expression is associated closely with the occurrence and development of cancers.25 Thus, firstly, we detected the expression of HNF1A-AS1 in TNBC cell lines (MDA-MB-453, MDA-MB-468, MDA-MB-231, BT-20 and MDA-MB-436) relative to normal human breast epithelial cell line (MCF-12F). The qRT-PCR results indicated that HNF1A-AS1 was upregulated in TNBC cells, particularly in BT-20 and MDA-MB-436 cells (Figure 1A). Besides, we also wondered its expression pattern in breast cancer cell lines originated from other subtypes. Interestingly, HNF1A-AS1 expression seemed to be also elevated in ER-positive T47D and MCF7 cells but almost unchanged in HER-positive MDA-MB-453 and SKBR3 cells (Figure S1A). Thereafter, the clinical data further validated the upregulation of HNF1A-AS1 in 28 TNBC samples compared with the paired non-tumor controls (Figure S1B). Thus, we speculated that HNF1A-AS1 might have some relation to TNBC progression. To certify this, we then investigated the biological function of HNF1A-AS1 in BT-20 and MDA-MB-436 cells. Through utilizing shRNAs targeting HNF1A-AS1, we discovered that HNF1A-AS1 expression was knocked down in BT-20 and MDA-MB-436 cells (Figure 1B). After that, we designed and conducted loss-of-function experiments in BT-20 and MDA-MB-436 cells. The results from colony formation assay manifested that the number of colonies was declined after silencing HNF1A-AS1 (Figure 1C), indicating that the proliferative capability of BT-20 and MDA-MB-436 cells was restrained by HNF1A-AS1 depletion. Meanwhile, EdU staining experiment also verified that cell proliferation could be hampered by silencing HNF1A-AS1, validated by the notable reduction of EdU positive cells in this case (Figures 1D and S1C). In the meantime, we evaluated the influence of silencing HNF1A-AS1 on cell apoptosis via JC-1 assay, TUNEL assay and flow cytometry analysis. It was discovered that JC-1 ratio was decreased when HNF1A-AS1 was knocked down in both TNBC cells (Figure 1E). Also, the proportion of TUNEL positive cells was elevated after HNF1A-AS1 was inhibited (Figures 1F and S1D). Moreover, cell apoptosis rate was obviously heightened after silencing HNF1A-AS1 (Figure 1G). These results demonstrated that cell apoptosis could be enhanced by HNF1A-AS1 deficiency. To further verify the role of HNF1A-AS1 in TNBC tumorigenesis, we also carried out in vivo xenograft experiments. As anticipated, in comparison to the data obtained from the control group, loss of HNF1A-AS1 slowed tumor growth rate (Figure S2A), resulting in smaller tumor size and lighter tumor weight (Figure S2B–C). Therefore, we concluded that HNF1A-AS1 was upregulated in TNBC, and silencing HNF1A-AS1 could repress the tumorigenesis of TNBC in vitro and in vivo.
HNF1A-AS1 Was Transcriptionally Stimulated by GATA1 in TNBC Cells
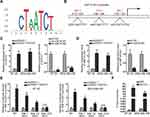
Reports have revealed the crucial role of transcription factors (TFs) in the dysregulation of lncRNAs in cancers.26 Here, we adopted UCSC (University of California, Santa Cruz; http://genome.ucsc.edu/) and JASPAR (http://jaspar.genereg.net/) to discover the probable TFs of HNF1A-AS1. As a result, GATA-binding protein 1 (GATA1) was found to potentially al regulate HNF1A-AS1 transcription. In addition, according to the binding motif of GATA1 predicted by JASPAR (Figure 2A), we discovered three latent binding sites for GATA1 in HNF1A-AS1 promoter (Figure 2B). After that, we validated that the expression of GATA1 was boosted by pcDNA3.1/GATA1 transfection but reduced by the transfection of sh-GATA1#1/2 in BT-20 and MDA-MB-436 cells through qRT-PCR (Figure 2C). Importantly, qRT-PCR tested that HNF1A-AS1 expression in BT-20 and MDA-MB-436 cells was strengthened under GATA1 overexpression but declined by GATA1 depletion (Figure 2D), indicating a positive correlation between GATA1 and HNF1A-AS1 in TNBC. Then, luciferase reporter experiment was implemented and the results demonstrated that the luciferase activity of wild-type HNF1A-AS1 promoter was visibly elevated by GATA1 overexpression, while such elevation could be gradually counteracted after mutation in the predicted three sites and completely offset when all the sites were mutated (Figure 2E). This suggested that GATA1 could bind with HNF1A-AS1 promoter at all the three sites. Moreover, ChIP assay data further proved that HNF1A-AS1 promoter was highly enriched in anti-GATA1-precipitated complexes (Figure 2F). Taken together, we confirmed that HNF1A-AS1 was transcriptionally activated by GATA1 in TNBC.
HNF1A-AS1 Sponged miR-32-5p in TNBC Cells
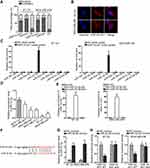
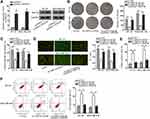
Researches have indicated that the regulatory mechanism lncRNAs exerted in cancer development depended on their localization in cancer cells.27 Hence, we implemented subcellular fractionation assay before investigating the regulatory mechanism of HNF1A-AS1 in TNBC. The results displayed that HNF1A-AS1 was mostly located in the cytoplasm of BT-20 and MDA-MB-436 cells (Figure 3A). Subsequently, the results of FISH assay further corroborated that HNF1A-AS1 was a cytoplasmic lncRNA in TNBC cells (Figure 3B). Recently, lncRNAs have been recognized to serve as a miRNA sponge to release miRNA-targeted mRNAs in cytoplasm.16 Thus, we utilized ENCORI (The Encyclopedia of RNA Interactomes; http://starbase.sysu.edu.cn/) to search for the miRNAs binding to HNF1A-AS1, and then eight miRNAs were discovered. Subsequently, RNA pull-down experiments were carried out to screen out the most suitable miRNA in TNBC cells. The results indicated that miR-32-5p could be visibly enriched by HNF1A-AS1 biotin probe, while other miRNAs did not represent notable enrichments (Figure 3C). Based on this, we selected miR-32-5p as the downstream miRNA of HNF1A-AS1 and tested its expression in TNBC cell lines through qRT-PCR. Results displayed a significant reduction of miR-32-5p expression in TNBC cells (Figure 3D). After that, RNA pull-down experiment further indicated that HNF1A-AS1 could be enriched by biotinylated miR-32-5p-WT rather than biotinylated miR-32-5p-Mut (Figure 3E), which demonstrated that miR-32-5p could bind with HNF1A-AS1. Then, we predicted the binding site of miR-32-5p in HNF1A-AS1 through ENCORI (Figure 3F). After overexpressing miR-32-5p in BT-20 and MDA-MB-436 cells with the transfection of miR-32-5p mimics (Figure 3G), we conducted luciferase reporter experiments to further verify the correlation between miR-32-5p and HNF1A-AS1. As a result, we discovered that the luciferase activity of HNF1A-AS1-WT was reduced by upregulated miR-32-5p, while that of HNF1A-AS1-Mut did not be affected under such circumstance (Figure 3H). In conclusion, HNF1A-AS1 could sponge miR-32-5p in TNBC cells.
MiR-32-5p Restrained Cell Proliferation and Facilitated Cell Apoptosis in TNBC
Then, we continued to search the function of miR-32-5p in TNBC cells. Seen from the data of colony formation assay, we discovered a remarkable reduction in the number of colonies under miR-32-5p overexpression (Figure 4A). Also, the outcomes of EdU assay also displayed that upregulating miR-32-5p led to an obvious decrease in the percent of EdU positive cells (Figure 4B). These data indicated that cell proliferation could be inhibited by miR-32-5p overexpression. Then, cell apoptosis was estimated through JC-1, TUNEL and flow cytometry experiments. The results of JC-1 experiment displayed that upregulating miR-32-5p evidently lessened the JC-1 ration in the two kinds of TNBC cells (Figure 4C). Also, the rate of TUNEL positive cells was increased due to miR-32-5p upregulation in BT-20 and MDA-MB-436 cells (Figure 4D). In addition, the apoptosis rates of both TNBC cells were elevated after overexpressing miR-32-5p (Figure 4E). These results demonstrated that cell apoptosis was accelerated by miR-32-5p overexpression. In short, we confirmed that miR-32-5p could restrain cell growth in TNBC.
RNF38 Was a Target of miR-32-5p in TNBC
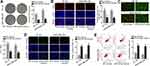
Subsequently, we began to search for the target mRNAs of miR-32-5p through ENCORI and 3 mRNAs were found under the shared prediction of PITA, RNA22, miRmap, micrpT and miRanda programs (Figure 5A). Then, qRT-PCR examined the expression of NSMAF (neutral sphingomyelinase activation associated factor), KLF4 (Kruppel like factor 4) and RNF38 (ring finger protein 38) in BT-20 and MDA-MB-436 cells transfected with miR-32-5p mimics. Intriguingly, ectopic expression of miR-32-5p resulted in a visible declination of RNF38 expression while had almost no impacts on the levels of other mRNAs (Figure 5B). Thus, we selected RNF38 to conduct further experiments. The results of qRT-PCR analysis displayed that RNF38 was highly expressed in TNBC cells (Figure 5C). Then, we utilized ENCORI to predict the binding site between RNF38 and miR-32-5p (Figure 5D). Thereafter, RIP assay was conducted and the results indicated that HNF1A-AS1, RNF38 and miR-32-5p were all enriched in the anti-Ago2 group (Figure 5E), demonstrating that all of them existed in RNA-induced silencing complex (RISC). Next, RNA pull-down experiments were implemented and the results indicated that RNF38 could be strongly enriched by biotinylated miR-32-5p-WT (Figure 5F). Furthermore, based on the data of luciferase reporter experiments, we discovered that only the luciferase activity of RNF38 3ʹUTR-WT was declined by miR-32-5p overexpression (Figure 5G). All in all, we verified that RNF38 was a target of miR-32-5p in TNBC.
HNF1A-AS1 Accelerated the Progression of TNBC via Regulating RNF38
To inquire whether HNF1A-AS1 could affect TNBC progression by regulating RNF38, we implemented rescue experiments. Before that, the overexpression efficiency of RNF38 in BT-20 and MDA-MB-436 cells was determined by qRT-PCR and Western blot experiments. The results displayed that both the mRNA level and protein level of RNF38 were elevated under the transfection of pcDNA3.1/RNF38 (Figure 6A). Then, according to colony formation assay data, we observed the decreased number of colonies caused by silenced HNF1A-AS1 was then recovered by overexpressed RNF38 (Figure 6B). Besides, it was indicated through EdU assay that the percentage of EdU positive cells was declined owing to the absence of HNF1A-AS1, but this trend was reversed after the co-transfection of pcDNA3.1/RNF38 (Figures 6C and S2D). These results demonstrated that RNF38 overexpression restored the repressed cell proliferation caused by HNF1A-AS1 inhibition. Later, through JC-1 assay, we discovered that HNF1A-AS1 depletion exerted the repressive function in JC-1 ratio and RNF38 overexpression reversed this phenomenon (Figure 6D). Next, TUNEL assay displayed that the declined rate of TUNEL positive cells due to HNF1A-AS1 deficiency was then normalized after upregulating RNF38 (Figures 6E and S2E). Finally, through flow cytometry experiment, we discovered that cell apoptosis rate that was enhanced by HNF1A-AS1 depletion could be abolished with the co-transfection of pcDNA3.1/RNF38 (Figure 6F). Taken together, HNF1A-AS1 could accelerate the progression of TNBC via an RNF38-mediated manner.
Discussion
Triple-negative breast cancer (TNBC) is a highly invasive subtype of breast cancer with a high mortality rate and has become a major threat to the health of women worldwide. In recent years, an increasing number of researches have confirmed that lncRNAs exert significant regulatory functions in human cancers, including TNBC.28 For example, Tang et al uncovered that PVT1 could modulate the progression of TNBC through KLF5/beta-catenin signaling.29 Wang et al identified that C-MYC-induced SNHG12 facilitated cell proliferation and migration in TNBC.30 Moreover, MALAT1 was discovered to facilitate TNBC cell proliferation and invasion via targeting miR-129-5p.31 Here, we chose HNF1A-AS1 to be our research object since it was confirmed to be an oncogene in bladder cancer, non-small cell lung cancer, colorectal cancer and so on.12–14 In our research, we first discovered that HNF1A-AS1 was overexpressed in TNBC tissues and cell lines. Of interest, its upregulation could also be seen in ER-positive cell lines but not in HER2-positive cell lines. This might suggest the potential role of HNF1A-AS1 in ER-positive breast cancer as well, while such speculation needs to be validated in the future. Furthermore, we demonstrated that knockdown of HNF1A-AS1 restrained cell growth in vitro and hindered tumorigenesis in vivo in TNBC. All the data validated the oncogenic property of HNF1A-AS1 in TNBC, the similar function as it exerted in other cancer types like osteosarcoma32 and gastric cancer.33
Interestingly, we found GATA1 was the transcription factor (TF) of HNF1A-AS1. Previously, GATA1 has been unveiled to be overexpressed and also function as a TF in breast carcinomas.34,35 Through mechanism experiments, we proved the interaction of GATA1 with HNF1A-AS1 promoter and also discovered that HNF1A-AS1 expression could be promoted by GATA1. It indicated that HNF1A-AS1 could be transcriptionally activated by GATA1 in TNBC.
Previous reports indicated that lncRNAs could modulate cancer progression through the competing endogenous RNA (ceRNA) network in cytoplasm.36 It is worth noting that the ceRNA network is a main regulatory mechanism for lncRNAs localized in the cytoplasm to modulate gene expression at the post-transcriptional level.37 In our study, we first proved the predominant distribution of HNF1A-AS1 in the cytoplasm of TNBC cells. Then, we utilized bioinformatics tools and discovered miR-32-5p as the downstream miRNA of HNF1A-AS1 in TNBC. In others’ researches, miR-32-5p was found to restrain the malignant phenotypes in cervical cancer,20 clear cell renal cell carcinoma38 and even TNBC.39 Consistently, here we discovered the down-regulation of miR-32-5p in TNBC cells, and also unmasked that miR-32-5p overexpression repressed cell growth in TNBC.
Present work proved RNF38 as the target of miR-32-5p in TNBC. RNF38 was confirmed to be an oncogene in several cancers, such as gastric cancer,23 hepatocellular carcinoma,22 and non-small cell lung cancer.24 In our research, we discovered a remarkable upregulation of RNF38 in TNBC cells. Besides, RNF38 was positively regulated by HNF1A-AS1 and negatively modulated by miR-32-5p. Furthermore, RNF38 was proved to compete with HNF1A-AS1 for the binding of miR-32-5p. In the end, rescue experiments demonstrated that overexpressed RNF38 could offset the restraining influence of silencing HNF1A-AS1 on TNBC cell growth.
Present research unveiled the oncogenic role of HNF1A-AS1 in TNBC through in vitro and in vivo experiments, and this has also been unveiled by a recent report focusing on breast cancer.40 Intriguingly, although both the two studies suggested HNF1A-AS1 as a ceRNA, our work disclosed the HNF1A-AS1/miR-32-5p/RNF38 network in TNBC, while the abovementioned literature discovered HNF1A-AS1/miR-20a-5p/TRIM32 axis in breast cancer.40 Besides, the current research further figured out that HNF1A-AS1 upregulation in TNBC was transcriptionally induced by GATA1.
However, the lack of the data of the correlations among HNF1A-AS1, miR-32-5p and RNF38 in TNBC samples is still a shortcoming due to the limited sample size, so are the data of their association with the survival of TNBC patients. Also, the downstream mechanism whereby RNF38 contributes to TNBC progression is covered presently. Moreover, since we also found the distribution of some HNF1A-AS1 in the nucleus, we have to admit that it is one of our limitations for not exploring the nuclear role of HNF1A-AS1 in TNBC cells. Importantly, we believe all the limitations will be overcome in future studies.
Conclusion
Taken together, GATA1-activated HNF1A-AS1 exerted the carcinogenic effect on TNBC. In addition, we proved that HNF1A-AS1 could facilitate the progression of TNBC via sponging miR-32-5p to upregulate RNF38. This discovery may provide a new idea to explore the therapeutic strategies of TNBC.
Data Sharing Statement
Data have all been provided in the manuscript and supplementary materials.
Ethics Approval and Informed Consent
This work was carried out with the approval of the Ethics Committee of the First People’s Hospital of Yunnan Province. All involvers signed the informed consents before surgery.
Consent for Publication
All involvers signed the informed consents before surgery.
Acknowledgment
We thank all the participators.
Funding
There is no funding to report.
Disclosure
No conflicts of financial interest are enclosed in this study.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Li N, Deng Y, Zhou L, et al. Global burden of breast cancer and attributable risk factors in 195 countries and territories, from 1990 to 2017: results from the Global Burden of Disease Study 2017. J Hematol Oncol. 2019;12(1):140. doi:10.1186/s13045-019-0828-0
3. Lin Y, Fu F, Lin S, et al. A nomogram prediction for the survival of patients with triple negative breast cancer. Oncotarget. 2018;9(63):32108–32118. doi:10.18632/oncotarget.24964
4. Wang C, Kar S, Lai X, et al. Triple negative breast cancer in Asia: an insider’s view. Cancer Treat Rev. 2018;62:29–38. doi:10.1016/j.ctrv.2017.10.014
5. Karaayvaz M, Cristea S, Gillespie SM, et al. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun. 2018;9(1):3588. doi:10.1038/s41467-018-06052-0
6. King JL, Zhang B, Li Y, et al. TTK promotes mesenchymal signaling via multiple mechanisms in triple negative breast cancer. Oncogenesis. 2018;7(9):69. doi:10.1038/s41389-018-0077-z
7. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
8. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
9. Lingling J, Xiangao J, Guiqing H, Jichan S, Feifei S, Haiyan Z. SNHG20 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through acting as a miR-154 sponge. Biomed Pharmacother. 2019;112:108648. doi:10.1016/j.biopha.2019.108648
10. Yang L, Xue Y, Liu J, et al. Long noncoding RNA ASAP1-IT1 promotes cancer stemness and predicts a poor prognosis in patients with bladder cancer. Neoplasma. 2017;64(6):847–855. doi:10.4149/neo_2017_606
11. Zhao X, Liu Y, Yu S. Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochim Biophys Acta Mol Basis Dis. 2017;1863(7):1805–1816. doi:10.1016/j.bbadis.2017.04.014
12. Feng Z, Wang B. Long non-coding RNA HNF1A-AS1 promotes cell viability and migration in human bladder cancer. Oncol Lett. 2018;15(4):4535–4540. doi:10.3892/ol.2018.7878
13. Zhang G, An X, Zhao H, Zhang Q, Zhao H. Long non-coding RNA HNF1A-AS1 promotes cell proliferation and invasion via regulating miR-17-5p in non-small cell lung cancer. Biomed Pharmacother. 2018;98:594–599. doi:10.1016/j.biopha.2017.12.080
14. Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F. Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/beta-catenin signaling pathway. Biomed Pharmacother. 2017;96:877–883. doi:10.1016/j.biopha.2017.10.033
15. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286.
16. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
17. Liu L, Chen Y, Li Q, Duan P. lncRNA HNF1A-AS1 modulates non-small cell lung cancer progression by targeting miR-149-5p/Cdk6. J Cell Biochem. 2019;120(11):18736–18750. doi:10.1002/jcb.29186
18. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–154. doi:10.1016/S1672-0229(08)60044-3
19. Liang H, Tang Y, Zhang H, Zhang C. MiR-32-5p regulates radiosensitization, migration and invasion of colorectal cancer cells by targeting TOB1 gene. Onco Targets Ther. 2019;12:9651–9661. doi:10.2147/OTT.S228995
20. Liu YJ, Zhou HG, Chen LH, et al. MiR-32-5p regulates the proliferation and metastasis of cervical cancer cells by targeting HOXB8. Eur Rev Med Pharmacol Sci. 2019;23(1):87–95. doi:10.26355/eurrev_201901_16752
21. Paul S. Integration of miRNA and mRNA expression data for understanding etiology of gynecologic cancers. Methods Mol Biol. 2019;1912:323–338.
22. Peng R, Zhang PF, Yang X, et al. Overexpression of RNF38 facilitates TGF-beta signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):113. doi:10.1186/s13046-019-1113-3
23. Zhang J, Wu H, Yi B, et al. RING finger protein 38 induces gastric cancer cell growth by decreasing the stability of the protein tyrosine phosphatase SHP-1. FEBS Lett. 2018;592(18):3092–3100. doi:10.1002/1873-3468.13225
24. Xiong D, Zhu SQ, Wu YB, et al. Ring finger protein 38 promote non-small cell lung cancer progression by endowing cell EMT phenotype. J Cancer. 2018;9(5):841–850. doi:10.7150/jca.23138
25. Charles Richard JL, Eichhorn PJA. Platforms for investigating LncRNA functions. SLAS Technol. 2018;23(6):493–506. doi:10.1177/2472630318780639
26. Lambert M, Jambon S, Depauw S, David-Cordonnier MH. Targeting transcription factors for cancer treatment. Molecules (Basel, Switzerland). 2018;23(6):1479. doi:10.3390/molecules23061479
27. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15(3):177–186. doi:10.1016/j.gpb.2016.12.005
28. Tian T, Gong Z, Wang M, et al. Identification of long non-coding RNA signatures in triple-negative breast cancer. Cancer Cell Int. 2018;18:103. doi:10.1186/s12935-018-0598-8
29. Tang J, Li Y, Sang Y, et al. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37(34):4723–4734. doi:10.1038/s41388-018-0310-4
30. Wang O, Yang F, Liu Y, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. 2017;9(2):533–545.
31. Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother. 2017;95:922–928. doi:10.1016/j.biopha.2017.09.005
32. Cai L, Lv J, Zhang Y, Li J, Wang Y, Yang H. The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J Cell Mol Med. 2017;21(11):2654–2662. doi:10.1111/jcmm.12944
33. Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res. 2018;78(20):5877–5890. doi:10.1158/0008-5472.CAN-18-1011
34. Boidot R, Végran F, Jacob D, et al. The transcription factor GATA-1 is overexpressed in breast carcinomas and contributes to survivin upregulation via a promoter polymorphism. Oncogene. 2010;29(17):2577–2584. doi:10.1038/onc.2009.525
35. Seo MJ, Liu X, Chang M, Park JH. GATA-binding protein 1 is a novel transcription regulator of peroxiredoxin 5 in human breast cancer cells. Int J Oncol. 2012;40(3):655–664. doi:10.3892/ijo.2011.1236
36. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
37. Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53(3):231–245. doi:10.1080/10409238.2018.1447542
38. Wang M, Sun Y, Xu J, et al. Preclinical studies using miR-32-5p to suppress clear cell renal cell carcinoma metastasis via altering the miR-32-5p/TR4/HGF/Met signaling. Int J Cancer. 2018;143(1):100–112. doi:10.1002/ijc.31289
39. Wang R, Huang Z, Qian C, et al. LncRNA WEE2-AS1 promotes proliferation and inhibits apoptosis in triple negative breast cancer cells via regulating miR-32-5p/TOB1 axis. Biochem Biophys Res Commun. 2020;526(4):1005–1012. doi:10.1016/j.bbrc.2020.01.170
40. Meng Q, Wang L, Lv Y, Wu J, Shi W. Deletion of HNF1A-AS1 suppresses the malignant phenotypes of breast cancer cells in vitro and in vivo through targeting miRNA-20a-5p/TRIM32 axis. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2019.3168
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.