Back to Journals » OncoTargets and Therapy » Volume 16
Galectin-9 Expression is Correlated to Cervical Squamous Cell Carcinoma Progression and Overall Survival
Authors Mendieta-Carmona V , Delgado-López G, Reyes-Leyva J , Gutiérrez-Quiroz CT, Vazquez-Zamora VJ, Picazo-Mendoza DA, Montiel-Jarquín AJ, Martinez-Morales LP, Vallejo-Ruiz V
Received 3 August 2023
Accepted for publication 22 October 2023
Published 31 October 2023 Volume 2023:16 Pages 891—904
DOI https://doi.org/10.2147/OTT.S433710
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr John Maher
Victoriano Mendieta-Carmona,1,2 Guadalupe Delgado-López,2 Julio Reyes-Leyva,3 Claudia Teresita Gutiérrez-Quiroz,4 Víctor Javier Vazquez-Zamora,4 Denisse Alejandra Picazo-Mendoza,4 Alvaro José Montiel-Jarquín,4 Laura Patricia Martinez-Morales,5 Verónica Vallejo-Ruiz2
1Posgrado en Ciencias Biológicas, Universidad Nacional Autónoma de México, Ciudad de Mexico, Mexico; Posgrado en Ciencias Biológicas, Universidad NacionalAutónoma de México (Posgrado en Ciencias Biológicas, Unidad de Posgrado, Edificio D, 1 Piso, Circuito de Posgrados, Ciudad Universitaria, Coyoacán, CDMX, C.P. 04510, México; 2Centro de Investigación Biomédica de Oriente, Instituto Mexicano del Seguro Social, Atlixco, Puebla, México; 3Facultad de Ciencias Químicas, Benemérita Universidad Autónoma de Puebla, Puebla, México; 4Hospital de Especialidades, General Manuel Ávila Camacho, Instituto Mexicano Del Seguro Social, Puebla, México; 5CONAHCYT-CIBIOR, Atlixco, Puebla, México
Correspondence: Verónica Vallejo-Ruiz, Centro de Investigación Biomédica de Oriente, Instituto Mexicano del Seguro Social, Km 4.5 Carretera Federal Atlixco-Metepec, s/n, Atlixco, Puebla, Z.C. 74360, México, Tel +52 24 44 440 122, Email [email protected]; [email protected]
Purpose: To determine whether galectin-9 gene (LGALS9) expression is correlated with cervical cancer progression, clinicopathological characteristics, and overall survival. To determine the biological processes and the abundance of tumour infiltrating immune cells related to the expression of LGALS9.
Patients and Methods: The study was conducted in two phases: 1) The expression level of LGALS9 was determined using the data of 193 squamous cell carcinoma (SCC) samples from The Cancer Genome Atlas (TCGA) database. Biological processes and tumour infiltrating cells associated to LGALS9 expression were evaluated using gene set enrichment analysis (GSEA) and tumour immune estimation resource (TIMER). 2) Independently, galectin-9 was identified in 40 SCC samples by immunohistochemistry and optical density quantified using ImagePro® software.
Results: The LGALS9 gene showed increased expression in cervical cancer samples. A higher expression level in SCC was related to better overall survival and to early clinical stages. GSEA showed that tumours with higher expression of LGALS9 were enriched in immune pathways such as interferon_alpha_response, and complement, the analysis of TIMER database showed a positive correlation between the expression level of LGALS9 and the abundance of tumour infiltrating immune cells. In addition, higher expression of galectin-9 was found in biopsies of SCC patients at early clinical stages, showing a trend of better survival.
Conclusion: Higher expression levels of LGALS9 and galectin-9 in SCC were related to early clinical stages and better prognosis. GSEA and TIMER analysis suggested that galectin-9 could play an antitumor role in cervical SCC.
Keywords: biomarker, galectin, cervical neoplasia, immune response, clinical stage, overall survival
Introduction
Cervical cancer is the fourth most common cancer in women worldwide, and the incidence and mortality rates differ drastically between low-income and high-income countries. Most cases occur in low-income countries, where many women are diagnosed in advanced stages, leading to higher mortality rates.1
Lack of opportune diagnosis and effective medical treatment are the main causes of high mortality due to cervical cancer. Therefore, it is still necessary to find new therapeutic options that improve the survival of women with cervical cancer.2 Immune cells play important roles in the vigilance and elimination of malignant cells, but evading tumour cells secret an array of soluble factors including cytokines, chemokines and growth factors that modify surrounding normal and immune cells to promote an immunosuppressive state that favour cancer development. Tumour cells act on immune cells by inducing several regulatory pathways including, but not limited to, checkpoint molecules, a group of proteins that control the timelapse and intensity of immune response, such as cytotoxic T-lymphocyte protein 4 (CTL4), programmed cell death protein 1 (PD-1) and T cell immunoglobulin and mucin domain-3 (TIM-3). Current cancer therapies are directed against checkpoint proteins to restore the protective role of immune cells.3,4
Galectins constitute a family of glycan-binding proteins that play central roles in the reprogramming of immune cells in the tumour microenvironment (TME). There have been recognized at least 15 Galectins (Gal-1 to Gal-15) that share the capacity to interact with β-galactoside-containing glycoconjugates but differ in their fine specificity and their biological activities. Galectins may behave as pro-tumoral (Gal-1) or antitumoral factors (Gal-3) and can be up- or down-regulated during cancer development.5,6
One of the most intriguing members of the group is Galectin-9, it is expressed in different epithelial tumours, including cervical carcinoma.7–10 Galectin-9 is a tandem repeat protein with two nonhomologous carbohydrate recognition domains that bind to different oligosaccharide structures.11 Galectin-9 can play several roles depending on its interaction with different ligands and can promote or inhibit tumour activity.12 It has been shown that galectin-9 participates in tumour cell adhesion, aggregation, migration, apoptosis, and growth.9,13,14
TIM-3 is the main receptor of Galectin-9, it is expressed on the membrane of both immune and tumour cells. The interaction of TIM-3 with galectin-9 induces apoptosis of Th-1 cells and is involved in the evasion capability of cancer cells.15–17
Galectin-9 may exert antitumour effects in different cancer types. In breast cancer, galectin-9 induces tumour cell aggregation and prevents metastasis, these effects correlated with increased survival.18,19 In colorectal cancer, galectin-9 induces apoptosis and suppresses proliferation;20 consequently, decreased expression of galectin-9 has been associated with poor outcome.21
Previous studies showed high galectin-9 expression in cervical cancer patients without lymph node metastasis, showing a trend towards improved survival.10,22 Contradictorily, cervical cancer patients positive for human papillomavirus showed higher levels of TIM-3 and galectin-9 that promote Treg cell differentiation that inhibits the cytotoxic activity of CD8+ T cells.23
The roles of galectin-9 in tumour cells suggest that galectin-9 could be a potential therapeutic target for the treatment of cervical cancer. Therefore, studying the expression of LGALS9 and the coding protein galectin-9 in the progression of cervical cancer will provide valuable information on its potential use as a therapeutic molecule.
The purpose of this work was to evaluate the association of galectin-9 expression with clinicopathological characteristics, and overall survival in cervical cancer patients. To identify the biological processes and the abundance of tumour infiltrating immune cells related to the expression of LGALS9.
Materials and Methods
Database Analysis
Data were obtained from the TCGA TARGET GTEx study included in the XENA browser (https://xenabrowser.net/). As a preliminary analysis, the RNAseq database was utilized to compare the mRNA expression levels of the galectins LGALS1, LGALS2, LGALS3, LGALS4, LGALS7, LGALS8, LGALS9, LGALS12, LGALS13 and LGALS14 between normal cervical tissue (n = 10) and squamous cell carcinoma (SCC), (n = 193) or adenocarcinoma (AC), (n = 37). The expression levels of mRNA were evaluated to determine if they were associated with overall survival. Because only LGALS9 expression was associated with overall survival in SCC, the study focused on LGALS9 expression in SCC and its relationship with clinicopathological characteristics. For this, a selection of samples from TCGA Cervical Cancer (CESC) was analysed considering the following criteria: primary site, sample type, and histological type, this analysis only included SCC samples (n = 183) (Table 1).
 |
Table 1 Characteristics of Patients with Cervical SCC Included in the RNAseq Analysis and in the Immunohistochemistry Assay |
To determine the biological processes related to the expression of LGALS9, a gene set enrichment analysis (GSEA) was performed, with samples grouped into high or low mRNA levels according to median expression. The Molecular Signatures Database v2023.1 was used to select hallmark gene sets. The number of permutations performed was 1000. A NOM p value < 0.05 and an adjusted q-value FDR < 0.25 were considered significant.
Additionally, to evaluate the correlation between the mRNA of the LGALS9 gene and tumour infiltrating immune cells, we analysed the data included data of the Tumor Immune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/).
Patients and Samples
The study was conducted in accordance with the Declaration of Helsinki and the ethical regulations approved by the Human Ethics Committee 785 from the Mexican Institute of Social Security (IMSS), registration number (R-2017-785-119).
The female patients included in the study were invited to participate at the Radiotherapy Service of the Unidad Médica de Alta Especialidad-Hospital de Especialidades (UMAE-HE), IMSS, at Puebla City, Mexico, during the period February 2017 to February 2022. All patients included in the study signed the informed consent form. The clinicopathological characteristics were obtained from clinical records. Patients included in the study did not receive prior treatment.
Galectin-9 Detection in Cervical Cancer Tissue
Paraffin-embedded biopsies of SCC of the cervix were obtained from the Department of Pathology of the UMAE-HE, IMSS. A total of 40 biopsies were included in the study, and the clinicopathological characteristics were obtained (Table 1). Histological Sections 4 µm thick were cut and placed on slides treated with (3-aminopropyl) triethoxysilane (APES) (440,140 Sigma Aldrich, Saint Louis, MO, USA); next, they were deparaffinized and rehydrated. Afterwards, antigen retrieval was conducted by incubation with Tris-Buffer Saline for 15 min at 90°C. Every step was followed by washing three times with 1x PBS, except after incubation with blocking buffer. Sections were incubated with 0.3% H2O2 in 1x PBS for 15 min to block endogenous peroxidase activity. Next, the tissues were incubated with blocking buffer containing 5% bovine serum albumin (P6154, Biowest, Riverside, MO, US) in 1× PBS for 1 hour. Next, the sections were incubated at 4°C overnight with the anti-Galectin-9 primary antibody (54,330, Cell Signalling, Massachusetts, USA) at a dilution of 1:250 in PBS containing 1% bovine serum albumin.
Afterwards, the tissues were incubated for 1 hour with anti-rabbit IgG-HRP (ab6721, Abcam, Cambridge, UK) at a dilution of 1:1000. The colorimetric reaction was assessed with ImmPACT® DAB Peroxidase Substrate (SK-4105, Vector Labs, Burlingame, CA, US). Nuclei were counterstained with haematoxylin (HX87960674 Merck, Darmstadt, Hesse, DE), and the slides were mounted with VectaMount® Permanent Mounting Medium (H-5000, Vector Labs, Burlingame, CA, UU).
Immunostaining was scored with Image-Pro software (Media Cybernetics, Inc., Rockville, MD, US). For each sample, 5 photos were taken, and the staining density was determined in 5 areas of each photo. The results corresponded to the mean optical density of all values and was reported as galectin-9 protein expression levels. The galectin-9 expression level was evaluated to determine its relationship with the clinicopathological characteristics of the patients and overall survival.
Statistical Analysis
To compare the mRNA levels of LGALS genes between normal cervical tissue and cervical cancer tissue, a one-way ANOVA test of a factor or Kruskal‒Wallis test was used according to the normality of the data, along with a Bonferroni or Dunn’s multiple comparisons post-hoc test, respectively. To determine whether the mRNA and galectin-9 expression levels were related to the survival of the patients, Kaplan‒Meier curves and a log rank analysis were used.
Univariate and multivariate Cox regression analyses were performed to determine the association between LGALS9 expression and several variables, including clinical stage, pathological grading, keratinization, and overall survival.
To identify the enriched groups of genes in the samples with high levels of the LGALS9 gene, GSEA was performed with GSEA_4.3.2 software. To determine whether the mRNA and protein expression levels of LGALS9 were related to the clinicopathological features, Student’s t test or the Mann‒Whitney U-test was used depending on the normality of the data. A significance level of α <0.05 was used, and the statistical tests were performed in GraphPad Prism 10.0.0.
A Spearman correlation was performed for the abundance of tumour infiltrating immune cells and expression level of LGALS9 gene.
Results
RNA Levels of Galectin Genes in Cervical Cancer vs Normal Tissues
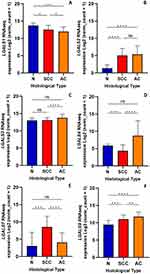
The changes in the mRNA levels of galectin genes (LGALS1, LGALS2, LGALS3, LGALS4, LGALS7, LGALS8, LGALS9, LGALS12, LGALS13 and LGALS14) in SCC and AC samples compared with normal tissue were determined using the RNAseq database from TCGA TARGET GTEx. LGALS1 expression showed a slight reduction in SCC and more evident in AC compared with normal tissues (Figure 1A). LGALS2 increased notably in both SCC and AC compared to normal samples (Figure 1B). LGALS4 expression decreased in SCC but increased in AC samples (Figure 1D). LGALS7 expression increased in SCC but did not differ in AC compared to normal tissues (Figure 1E). LGALS9 expression increased in SCC and AC (p = 0.0002 and p < 0.0001 respectively). Taken together, higher expression of LGALS3, LGALS4 and LGALS9 was found in AC than in SCC samples (Figure 1C, D and F), while LGALS1 and LGALS7 expression was higher in SCC than in AC. LGALS8 and LGALS12 expression did not show significant changes in cervical cancer (data not shown). LGALS13 was not detected in normal samples while LGALS14 was detected in a limited number of samples (data not shown); therefore, they were not further analysed.
RNA Levels of LGALS9 and Its Association with Overall Survival and Clinicopathological Characteristics
Further bioinformatic analysis were performed to identify the changes in LGALS genes expression associated to the overall survival. For this, LGALS genes that showed significant differences in mRNA levels in cervical cancer compared with normal tissues were divided into high and low expression groups, considering the median. Only for the LGALS9 gene it was observed a statistical significance, higher levels of the LGALS9 gene (p < 0.0001) were associated with better survival in patients with SCC (Figure 2A). There was no association between LGALS9 expression and survival in patients with AC (p = 0.9726; Figure 2B).
LGALS9 expression changes was also associated with clinicopathological characteristics in SCC; indeed, decreased LGALS9 expression was associated to advanced clinical stages III and IV (p = 0.0072; Figure 3). There were no differences in other pathological characteristics analysed, such as differentiation degree, keratinization, and lympho-vascular invasion (data not shown).
Univariate and multivariate Cox regression analyses were performed to determine the association between overall survival and the LGALS9 expression level, clinical stage, pathological grading, and keratinizing. Both the univariate and multivariate analyses showed a statistical association in the expression level of LGALS9 and the clinical stage showing that they were independent prognostic factors (Table 2).
 |
Table 2 Univariate and Multivariate Cox Regression Analysis of Clinical Stage, Pathological Grading, Keratinizing and LGALS9 Expression Level |
Enriched Pathways Associated to LGALS9 Expression
GSEA was performed in samples with high and low LGALS9 expression, and the analysis showed 7 pathways related to high LGALS9 expression (Table 3). Some of them are involved in the immune response, such as complement, interferon_alpha_response, and other pathways are involved in cellular metabolism, such as xenobiotic_metabolism and bile_acid_metabolism. The results showed some of the biological pathways enriched with the high expression of LGALS9 are implicated in the regulation of immune processes.
 |
Table 3 Gene Set Enrichment Analysis of High LGALS9 Expression in Squamous Cervical Carcinoma Compared with Low LGALS9 Expression |
LGALS9 Expression is Associated with Abundance of Tumour Infiltrating Immune Cells
The correlation between LGALS9 expression and the tumour infiltrating immune cells was determined with data of the TIMER database. Bioinformatic analysis showed that LGALS9 expression was correlated with several immune cell infiltration. The strongest positive correlation was observed in neutrophils (R = 0.406, p < 0.0001); but there were slight correlations in CD4+ T cells (R = 0.342, p < 0.0001), B cells (R = 0.286, p < 0.0001), CD8+ T cells (R = 0.229, p=<0.001) and dendritic cells infiltration (R = 0.198, p < 0.001), see Table 4.
 |
Table 4 Correlation Between the LGALS9 Expression Level and the Abundance of Immune Cells Infiltrates in Cervical and Endocervical Cancer Tumours, Based on TIMER Database |
Galectin-9 Identification in Squamous Cervical Cancer Tissue
Galectin‑9 expression level, the nuclear and cytoplasmic location in cervical cancer tissue varied between samples (Figure 4A and B). Some samples showed both cytoplasmic and nuclear expression (Figure 4C), others showed only a cytoplasmic location (Figure 4B), and some cellular nuclei showed high expression of galectin-9 (Figure 4D). The cellular localization was not related to the clinicopathological characteristics.
Based on the differences in the galectin-9 expression level between samples, we decided to determine whether the expression level was related to the clinicopathological characteristics. For the clinical stage characteristics, a significant decrease in galectin-9 expression was observed in advanced clinical stages (p = 0.0336) (Figure 5A). The analysis showed no relationship between the galectin-9 expression level and the degree of differentiation (G1/G2 vs G3/G4) and keratinization (p = 0.2865 and p = 0.0671, respectively) (Figure 5B and C).
The group of deceased patients showed lower galectin-9 expression in cancer tissue than the group of patients in remission of the disease (p = 0.0487), (Figure 5D).
Galectin-9 Expression Level in Cervical Squamous Cell Carcinoma and Its Association with Overall Survival
To determine if the galectin-9 expression level in SCC tissue was related to the overall survival of the patients, the samples were divided into two groups, high and low expression levels, considering the median of the group. After that, a Kaplan‒Meier curve and a log rank analysis were performed. The results did not show a significant relationship (p = 0.1378); however, it is important to note that there was a tendency of better survival for the patients with higher expression of galectin-9 (Figure 6).
Discussion
Galectins play many different roles in cancer biology depending on their cellular or extracellular location, concentration, and the interaction with specific ligands in normal and immune cells.24
Galectin genes can be up- or down-regulated in different cancer types, including cervical cancer, and have been involved in cancer progression.25
In this study, we performed bioinformatic analysis to evaluate the expression of LGALS genes from a public database of cervical cancer and normal samples. Our results identified changes in the mRNA levels of LGALS1, LGALS2, LGALS4, LGALS7 and LGALS9. Previous studies reported increased expression of Galectin-1 in cervical cancer tumours, but our results showed decreased levels of mRNA of LGALS1.10 LGALS4 showed decreased expression in squamous cell carcinoma. Galectin-4 was evaluated by our research group in SCC, the expression level was higher in keratinizing tumours than in nonkeratinizing tumours, and a trend of higher expression was observed in tumours of clinically advanced stages.26 The LGALS7 gene showed increased expression in squamous cell carcinoma, but studies of galectin-7 in cervical cancer reported decreased expression, finding that tumours with high expression were related to a better prognosis.27 Therefore, the mRNA levels of LGALS1, LGALS4 and LGALS7 genes was not related to protein expression, suggesting that a posttranscriptional mechanism could modulate protein synthesis.
Our results showed significant increased expression of LGALS9 in both SCC and AC. In addition, patients with SCC and higher LGALS9 expression showed a significant better survival, but not those with AC. Contrarily, Zhang et al, 2019, found that lower levels of LGALS9 expression were associated with better overall survival.28 However, their study included all the histological types in the correlation analysis; meanwhile our study analysed separately SCC and AC data, considering that they are tumours with different molecular characteristics.
In addition, we found association of higher expression of LGALS9 with early clinical stages of SCC. Our findings of mRNA levels are supported with the expression of galectin-9 protein, Punt et al reported that cervical cancer tumours positive to galectin-9 were related with better survival.10
Additionally, enrichment analysis was performed to identify the pathways related to high expression of LGALS9. The results showed that galectin-9 in cervical cancer tumours is associated to the immune response pathways, as complement, and interferon_alpha_response, highlighting its relevance in the modulation of the tumour microenvironment.
The bioinformatic analysis showed that LGALS9 expression was positively associated with immune cell infiltration, presenting from highest to lowest abundance: neutrophils, CD4+ T cells, B cells, CD8+ T cells and dendritic cells. Although this is an initial approach, the role of neutrophils in cancer development is of great interest because they are included as a diagnostic marker in several types of cancer; indeed, in ex vivo assays it has been reported that galectin-9 activate neutrophils, and also increase neutrophils’ life span,29 other studies reported that galectin-9 activate neutrophil-mediated cytotoxic activity in cancer cells.30 Additionally, it has been reported that galectin-9 could play an antitumor immunity activating antigen presentation by dendritic cells,31 although in our study dendritic cells infiltration showed the lowest correlation.
Galectin-9 expression was identified in both the nucleus and cytoplasm of tumour cells. Beyer reported galectin-9 location only in the cytoplasm of SCC and they found that higher cytoplasmic galectin-9 expression in tumour cells is related to better prognosis.22 In the samples analysed, we observed tumours with only a cytoplasmic location and others with both a nuclear and cytoplasmic location, but we did not found a relationship between the subcellular location and any clinicopathological characteristics.22 Changes in galectin subcellular location in cancer have been reported for other galectins. The expression of galectin-7 increases in oesophageal squamous cell carcinoma tumours with respect to normal tissue; but it was mainly observed in the nucleus of normal cells, meanwhile it was found in the cytoplasm, nuclei, and membranes of cancer cells.32 Galectin-3 also changed its localization in melanoma; indeed, higher expression of nuclear galectin-3 was associated to better survival than patients with lower nuclear galectin-3 expression.33 The subcellular location of galectin-9 in cervical cancer tumours must be explored in future studies to determine a possible relationship with its functions.
In our study, we found higher galectin-9 expression in early clinical stages. Our results agree with a previous report that also found higher expression of galectin-9 in SCC tumours with lower FIGO status. Beyer et al reported higher expression levels in SCC and FIGO stage I, and they also reported higher expression of galectin-9 in lower grading tumours.22
We also evaluated galectin-9 expression with respect to patients with disease remission vs deceased patients, and higher expression was observed in the first group. The analysis of overall survival and galectin-9 expression levels showed a trend towards better survival in patients with higher expression levels. Punt et al also reported a trend of better survival for patients with tumours with higher expression of galectin-9.10 This is the first study that identify in cervical cancer tumours with high expression of LGALS9 the enrichment of genes related to immune pathways and major abundance on tumour infiltrating immune cells. The potential role of galectin-9 in the regulation of immune response in cervical cancer should be analysed in more detail in further studies.
Conclusion
Higher mRNA levels of LGALS9 and galectin-9 correlated with early clinical stages and better survival in patients with cervical SCC. In addition, LGALS9 levels were positively associated with immunological pathways and infiltration of tumour immune cells, highlighting the role of galectin-9 as an immunomodulator and a potential prognostic biomarker.
Acknowledgments
This work was supported by the Consejo Nacional de Ciencia y Tecnología de México, S0008-2017-1 grant number 290068; CONACYT-Infraestructura, grant number 300379. This paper is part of the requirements for obtaining a doctoral degree at the Posgrado en Ciencias Biológicas, UNAM of VMC. We thank the Consejo Nacional de Ciencia y Tecnología de México for support through a graduate scholarship.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Mayadev JS, Ke G, Mahantshetty U, et al. Global challenges of radiotherapy for the treatment of locally advanced cervical cancer. Int J Gynecol Cancer. 2022;32(3):436–445. doi:10.1136/ijgc-2021-003001
3. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi:10.1038/nature21349
4. Yang R, Sun L, Li CF, et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat Commun. 2021;12(1):832. doi:10.1038/s41467-021-21099-2
5. Elola MT, Ferragut F, Méndez-Huergo SP, et al. Galectins: multitask signaling molecules linking fibroblast, endothelial and immune cell programs in the tumor microenvironment. Cell Immunol. 2018;333:34–45. doi:10.1016/j.cellimm.2018.03.008
6. Liu FT, Stowell SR. The role of galectins in immunity and infection. Nat Rev Immunol. 2023;23(8):479–494. doi:10.1038/s41577-022-00829-7
7. Zhou X, Sun L, Jing D, et al. Galectin-9 expression predicts favorable clinical outcome in solid tumors: a systematic review and meta-analysis. Front Physiol. 2018;9:452. doi:10.3389/fphys.2018.00452
8. Choi S, Kook MC, Kook MC, et al. Prognostic value of tumoral expression of galectin-9 in gastric cancer. Turkish J Gastroenterol. 2017;28(3):166–170. doi:10.5152/tjg.2017.16346
9. Zhang ZY, Dong JH, Chen YW, et al. Galectin-9 acts as a prognostic factor with antimetastatic potential in hepatocellular carcinoma. Asian Pac J Cancer Prev. 2012;13(6):2503–2509. doi:10.7314/APJCP.2012.13.6.2503
10. Punt S, Thijssen VL, Vrolijk J, et al. Galectin-1, −3 and −9 expression and clinical significance in squamous cervical cancer. PLoS One. 2015;10(6):e0129119. doi:10.1371/journal.pone.0129119
11. John S, Mishra R. Galectin-9: from cell biology to complex disease dynamics. J Biosci. 2016;41(3):507–534. doi:10.1007/s12038-016-9616-y
12. Thijssen VL, Heusschen R, Caers J, et al. Galectin expression in cancer diagnosis and prognosis: a systematic review. Biochim Biophys Acta. 2015;1855(2):235–247. doi:10.1016/j.bbcan.2015.03.003
13. Nobumoto A, Nagahara K, Oomizu S, et al. Galectin-9 suppresses tumor metastasis by blocking adhesion to endothelium and extracellular matrices. Glycobiology. 2008;18(9):735–744. doi:10.1093/glycob/cwn062
14. Heusschen R, Griffioen AW, Thijssen VL. Galectin-9 in tumor biology: a jack of multiple trades. Biochim Biophys Acta. 2013;1836(1):177–185. doi:10.1016/j.bbcan.2013.04.006
15. Zhu C, Anderson AC, Schubart A, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6(12):1245–1252. doi:10.1038/ni1271
16. Sánchez-Fueyo A, Tian J, Picarella D, et al. Tim-3 inhibits T helper type 1–mediated auto- and alloimmune responses and promotes immunological tolerance. Nat Immunol. 2003;4(11):1093–1101. doi:10.1038/ni987
17. Pang N, Alimu X, Chen R, et al. Activated galectin-9/TIM3 promotes Treg and suppresses Th1 effector function in chronic lymphocytic leukemia. FASEB J. 2021;35(7):e21556. doi:10.1096/fj.202100013R
18. Yasinska IM, Sakhnevych SS, Pavlova L, et al. The Tim-3-Galectin-9 pathway and Its regulatory mechanisms in human breast cancer. Front Immunol. 2019;10:1594. doi:10.3389/fimmu.2019.01594
19. Yamauchi A, Kontani K, Kihara M, et al. Galectin-9, a novel prognostic factor with antimetastatic potential in breast cancer. Breast J. 2006;12(5 Suppl 2):S196–200. doi:10.1111/j.1075-122X.2006.00334.x
20. Morishita A, Nomura K, Tani J, et al. Galectin‑9 suppresses the tumor growth of colon cancer in vitro and in vivo. Oncol Rep. 2021;45(6):105. doi:10.3892/or.2021.8056
21. Wang Y, Sun J, Ma C, et al. Reduced expression of galectin-9 contributes to a poor outcome in colon cancer by inhibiting NK cell chemotaxis partially through the Rho/ROCK1 signaling pathway. PLoS One. 2016;11(3):e0152599. doi:10.1371/journal.pone.0152599
22. Beyer S, Wehrmann M, Meister S, et al. Galectin-8 and −9 as prognostic factors for cervical cancer. Arch Gynecol Obstet. 2022;306(4):1211–1220. doi:10.1007/s00404-022-06449-9
23. Chen Z, Dong D, Zhu Y, et al. The role of Tim-3/Galectin-9 pathway in T-cell function and prognosis of patients with human papilloma virus-associated cervical carcinoma. FASEB J. 2021;35(3):e21401. doi:10.1096/fj.202000528RR
24. Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5(1):29–41. doi:10.1038/nrc1527
25. Li CH, Chang YC, Chan MH, et al. Galectins in cancer and the microenvironment: functional roles, therapeutic developments, and perspectives. Biomedicines. 2021;9(9):1159. doi:10.3390/biomedicines9091159
26. Conde-Rodríguez I, Delgado-López G, Armenta-Castro E, et al. Evaluation of serum levels and expression of galectin-4 in cervical cancer. BioMed Res Int. 2020;2020:1–9. doi:10.1155/2020/6756723
27. Higareda-Almaraz JC, Ruiz-Moreno JS, Klimentova J, et al. Systems-level effects of ectopic galectin-7 reconstitution in cervical cancer and its microenvironment. BMC Cancer. 2016;16(1):680. doi:10.1186/s12885-016-2700-8
28. Zhang L, Tian S, Pei M, et al. Crosstalk between histone modification and DNA methylation orchestrates the epigenetic regulation of the costimulatory factors, Tim‑3 and galectin‑9, in cervical cancer. Oncol Rep. 2019;42(6):2655–2669. doi:10.3892/or.2019.7388
29. Vega-Carrascal I, Bergin DA, McElvaney OJ, et al. Galectin-9 signaling through TIM-3 is involved in neutrophil-mediated Gram-negative bacterial killing: an effect abrogated within the cystic fibrosis lung. J Immunol. 2014;192(5):2418–2431. doi:10.4049/jimmunol.1300711
30. Ustyanovska Avtenyuk N, Choukrani G, Ammatuna E, et al. Galectin-9 triggers neutrophil-mediated anticancer immunity. Biomedicines. 2021;10(1):66. doi:10.3390/biomedicines10010066
31. Nagahara K, Arikawa T, Oomizu S, et al. Galectin-9 increases Tim-3+ dendritic cells and CD8+ T cells and enhances antitumor immunity via galectin-9-Tim-3 interactions. J Immunol. 2008;181(11):7660–7669. doi:10.4049/jimmunol.181.11.7660
32. Zhu X, Ding M, Yu ML, et al. Identification of galectin-7 as a potential biomarker for esophageal squamous cell carcinoma by proteomic analysis. BMC Cancer. 2010;10(1):290. doi:10.1186/1471-2407-10-290
33. Brown ER, Doig T, Anderson N, et al. Association of galectin-3 expression with melanoma progression and prognosis. Eur J Cancer. 2012;48(6):865–874. doi:10.1016/j.ejca.2011.09.003
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.