Back to Journals » OncoTargets and Therapy » Volume 14
Functions and Targets of miR-335 in Cancer
Authors Ye L, Wang F, Wu H, Yang H, Yang Y, Ma Y, Xue A, Zhu J, Chen M, Wang J, Zhang QA
Received 3 February 2021
Accepted for publication 28 April 2021
Published 20 May 2021 Volume 2021:14 Pages 3335—3349
DOI https://doi.org/10.2147/OTT.S305098
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Lingling Ye, Fen Wang, Hao Wu, Hui Yang, Yan Yang, Yajun Ma, Aili Xue, Jing Zhu, Meili Chen, Jinyan Wang, Quan an Zhang
Department of Oncology, The Affiliated Jiangning Hospital with Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China
Correspondence: Quan an Zhang
Department of Oncology, The Affiliated Jiangning Hospital with Nanjing Medical University, Nanjing, 210000, Jiangsu, People’s Republic of China
Email [email protected]
Abstract: MicroRNAs (miRNAs) are small non-coding RNAs (18∼ 25 nt in length) that act as master regulators of eukaryotic gene expression. They might play an oncogenic or tumor-suppressive role in multiple cancers. In recent decades, several studies have focused on the functions and mechanisms of miR-335 in cancer. The expression level of miR-335 in tissues and cells varies with cancer types, and miR-335 has been proposed as a potential biomarker for the prognosis of cancer. Besides, miR-335 may serve as an oncogene or tumor suppressor via regulating different targets or pathways in tumor initiation, development, and metastasis. Furthermore, miR-335 also influences tumor microenvironment and drug sensitivity. MiR-335 is regulated by various factors such as lncRNAs and microRNAs. In this review, we reveal the functions and targets of miR-335 in various cancers and its potential application as a possible biomarker in prognostic judgment and treatment of malignant tumors.
Keywords: miR-335 cancer, prognosis, tumor biology, regulation
Introduction
MicroRNAs (miRNAs) are small noncoding regulatory RNAs ranging in size from 18 to 25 nucleotides. MiRNAs regulate gene expression mainly through recognizing complementary sites in 3ʹ-untranslated regions (3′-UTR) of target messenger RNAs (mRNAs).1 As regulatory biomolecules, miRNAs do not entail completely complementary to induce the regulation of their target genes. Therefore, a single miRNA can regulate various genes, and one single gene can be targeted by different miRNAs.2 Hence, the identification of target genes is a complex process.3 MiRNAs are dysregulated in multiple cancers and recognized as oncogenes or tumor suppressors according to their functions. Overexpression of oncomiRs promote tumor progression by inhibiting tumor-suppressive genes, such as miR-21,4 miR-221/222,5 and miR-27a.6,7 In contrast, tumor-suppressive miRNAs suppress tumor development by inhibiting oncogenes. Some miRNAs are considered as tumor suppressors, including miR-145,8 the let-7 family,9 and miR-205.10 Besides, miRNAs are involved in many cellular processes, some of which are considered hallmarks of cancer, including cell proliferation,11–13 apoptosis,14,15 metastasis,16,17 and so on. In conclusion, accumulating evidence has indicated that miRNAs play important roles in cancer development.
Among numerous miRNAs, miR-335 has attracted widespread attention. Numerous studies show that miR-335 is dysregulated in many cancers, such as breast cancer, lung cancer, colorectal cancer, and ovarian cancer,18–21 and acts as an oncogene or tumor suppressor in many malignant tumors. The differential expression of miR-335 in various cancers may indicate a differential relation between miR-335 expression and survival. Many pieces of research find the relevance between miR-335 expression and survival of cancer patients.22–24 Furthermore, the role of miR-335 in cancers depends on its influence on the biological behavior of multiple tumors, including proliferation,25 apoptosis,26 migration,27 and invasion.21 Additionally, more and more evidence has shown that miR-335 affects the response to chemotherapy or radiotherapy in cancer patients.24–28 Along with the production of miR-335, miR-335-3p (also known as miR-335*) is also produced. There is little research on miR-335-3p here. Zhao. W et al29 find that upregulated lncRNA CASC9 contributes to proliferation, migration, and invasion of Non-Small Cell Lung Cancer (NSCLC) through inhibition of miR-335-3p. And S100 calcium-binding protein A14 (S100A14) acts as a target of miR-335-3p.
This review discusses the pathways and targets of miR-335 involved in cancers and summarizes its functions as a biomarker for the prognosis. It will also describe novel results that miR-335 is proposed as a potential therapeutic target for cancers.
The Clinical Significance of miR-335 in Cancers
The Expression of miR-335 in Cancers
The Downregulation of miR-335 in Cancers
Growing research studies have studied the expression profile of miR-335 in cancers. The results of some research studies indicate that miR-335 is down-regulated in some cancers (Table 1). In detail, the expression of miR-335 is down-regulated in NSCLC, which is the commonest type of pulmonary malignant tumor.19,22,25,30,31 The downregulation of miR-335 is noticed in breast cancer, which is the most common malignancy in women.18,32–35 Likewise, in osteosarcoma, a tumor of the skeletal system, the expression level of miR-335 is reduced.27,36,37 MiR-335 is also decreased in esophageal squamous cell carcinoma (ESCC), gastric cancer (GC), hepatocellular carcinoma (HCC), colorectal carcinoma (CRC), and pancreatic cancer.23,26,38–43 The expression of miR-335 is decreased in various malignant tumors of the genitourinary system like ovarian cancer, renal cell carcinoma (RCC), bladder cancer, and prostate cancer.21,44–49 Additionally, the expression of miR-335 is down-regulated in thyroid cancer50 and melanoma tissues.24
 |
Table 1 Expression Patterns and Prognostic Value of miR-335 in Multiple Types of Human Cancer |
The Upregulation of miR-335 in Cancers
However, miR-335 is upregulated in several cancers. The expression of miR-335 is up-regulated in gallbladder carcinoma51 and endometrial cancer.52 Besides, the controversy over the expression of miR-335 in colorectal carcinoma (CRC) also still exists.20,43 As a result, the expression of miR-335 varies with types of cancers and might closely be relate to tumor heterogeneity. The controversial findings regarding miR-335 in some cancers require further study.
The Prognostic Value of miR-335 in Cancer
MiR-335 has been found to not only be dysregulated in many types of cancers but also function as a prognostic biomarker (Table 1). MiR-335 is usually decreased in GC. Among GC patients, the lower miR-335 expression is apparently correlated to higher clinical stage, venous invasion, poor differentiation, and poor OS.26,41 But Yan et al53 reported that a high level of miR-335 is associated with a high frequency of recurrence and poor survival rate. In NSCLC, the down-regulation of miR-335 is related to lymph node metastasis.22 The expression of miR-335 is negatively associated with the pathological stage and metastasis of CRC.54 Regarding ovarian cancer, low miR-335 is correlated to shorter OS and relapse-free survival (RFS) and emerged as an independent prognostic factor for OS and RFS. Compared with the nonrecurrence group, miR-335 is down-regulated in the recurrence group.45 In ESCC, it is proved that miR-335 expression is negatively associated with histological grade, lymph node metastasis, tumor stage. In addition, low miR-335 expression is correlated with poor prognosis in ESCC patients.23 In HCC, lower serum miR-335 levels are closely associated with higher mean serum AFP level, more vascular invasion, and larger tumor size. Patients with low serum miR-335 levels had a poor prognosis.55 Besides, the miR-335 expression is negatively associated with advanced stages of melanoma.24 Although miR-335 has been linked to the prognosis of multiple cancers, more studies are needed to confirm this common association.
Biological Role of miR-335 in Various Cancers
MiR-335 as a Tumor Suppressor miR
Proliferation and Apoptosis
Many diseases occur due to the disorder between cell proliferation and death, especially cancer. Emerging research studies have indicated that miR-335 affected proliferation and apoptosis in a variety of cancers (Table 2). Rho-associated coiled-coil containing protein kinase 1 (ROCK1), which is a serine/threonine kinase protein, facilitates the development of malignant tumors. In NSCLC, miR-335 modulates cell proliferation and cell cycle progression by down-regulating ROCK1expression.24 Besides, miR-335 suppresses the proliferation of cervical cancer cells and HCC cells by down-regulating ROCK1.56,57 In addition to regulating cell proliferation, miR-335 can also regulate migration and invasion in HCC.39 MiR-335 down-regulates the expression of ROCK1 through straightly binding to 3′-UTR of it, leading to reduced cell proliferation and migration in melanoma.24 In GC, miR-335 may play a part in proliferation inhibition by targeting ROCK1.58 Furthermore, miR-335 can be decoyed by lncRNA DANCR to facilitate ROCK1-mediated proliferation and migration in osteosarcoma.59
 |
Table 2 MiR-335 Regulates Cell Proliferation and Apoptosis in Various Cancer Types |
B-cell CLL/lymphoma 2 like 2 (BCL-W or BCL2L2) is also an oncoprotein involved in a variety of human tumors. BCL-W, which is an anti-apoptotic member of the Bcl-2 protein family, is identified as a possible target of miR-335 based on the results of bioinformatics analysis.21 In NSCLC, Bcl-w is identified as a target gene of miR-335. MiR-335 inhibits cell invasion and promotes apoptosis through targeting Bcl-w and SP1.19 In RCC, miR-335 inhibits proliferation and invasion by decreasing BCL-W.44,60 In ovarian cancer, the upregulation of miR-335 induces cell apoptosis, and BCL-W is a target of miR- 335.49
Apart from the pathways mentioned above, several signaling pathways are also involved in the proliferation and apoptosis of cancers. Cullin 4B (CUL4B) is a scaffold protein that belongs to the cullin four subfamily.61 MiR-335 suppresses the cell cycle process and promotes apoptosis by decreasing CUL4B in NSCLC.62 Transformer 2 beta homolog (Tra2β) is considered to be involved in the progression of several diseases, including cancer.63 MiR-335 inhibits cell proliferation of lung cancer cells by targeting Tra2β, which is up-regulated in NSCLC.25 In NSCLC, the highly expressed Copine 1 (CPNE1) promotes cell proliferation and is negatively regulated by miR-335.31 In addition, miR-335 inhibits proliferation and facilitates apoptosis via targeting superoxide dismutase 2 (SOD2), which is up-regulated in NSCLC.64 MiR-335 can inhibit cell proliferation, migration, and accelerate cell apoptosis in thyroid cancer by directly regulating Sonic Hedgehog SHH.65 High expression intercellular adhesion molecule 1 (ICAM-1) is also observed in thyroid cancer. MiR-335 inhibits proliferation and promotes apoptosis of thyroid cancer cells through controlling ICAM-1.50 In breast cancer, miR-335 impacts cell functions such as proliferation and apoptosis by regulating different targets in the upstream BRCA1-regulatory cascade.18 Survivin, which is an inhibitor of apoptosis protein, mainly regulates apoptosis and cell cycle control.66,67 In GC, miR-335 inhibits proliferation and enhances apoptosis via regulating survivin expression.26 Besides, miR-335 contributes to the increased apoptotic by derepression of survivin in osteosarcoma.37 MiR-335 functions as a tumor suppressor to modulate the proliferation of CRC cells by down-regulating lactate dehydrogenase B (LDHB).43 MiR-335 can inhibit proliferation and induce apoptosis by modulating the jumonji domain containing 2C (JMJD2C) in pancreatic cancer.68 In bladder cancer, miR-335 inhibits cell proliferation and migration through up-regulating of CT10 regulator of kinase-like protein (CRKL), which acts as an oncogene in many cancers.46 Furthermore, miR-335 can also inhibit proliferation by depressing octamer-binding transcription factor 4 (OCT4) in bladder cancer.48 OCT4, also named POU5F1, is the main regulator in maintaining the pluripotency of stem cells.69 In prostate cancer, early growth response 3 (EGR3) promotes cell proliferation and is negatively regulated by miR-335.47 In cervical cancer, miR-335 may inhibit cell proliferation by regulating protein phosphatase 6 catalytic subunit (PPP6C).70
Migration and Invasion
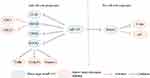
Numerous studies have shown that miR-335 modulates the expression of various genes, and plays important roles in the migration and invasion of cancer (Figure 1). ROCK1, a member of the ROCK family, is the downstream target of miR-335. In detail, in osteosarcoma, miR-335 acts as a tumor suppressor to inhibit cell migration and invasion by regulating the ROCK1.27 Similar results are reported in another two studies.59,71 MiR-335 regulates ROCK1 by binding with 3′-UTR of it diametrically and leads to the inhibition of migration in melanoma.24 In nasopharyngeal carcinoma, miR-335 inhibits invasion and metastasis of cancer cells by directly regulating ROCK1.72 Besides, miR-335 inhibits GC cell migration and invasion via regulating ROCK1.58 MiR-335 acts as a tumor suppressor to modulate cell migration and invasion by the down-regulation of ROCK1 in HCC.39 Li et.al57 reported the same results. Furthermore, through the suppression of ROCK1, miR-335 inhibits cell migration, invasion, and epithelial-mesenchymal transition (EMT) in cervical cancer and NSCLC.22,56
Apart from ROCK1, several genes are involved in the metastasis of cancers as well. MiR-335 facilitates cell invasion and migration by targeting ICAM-1 in thyroid cancer.50 Besides, miR-335 represses SHH and inhibits the migratory and invasive potential of thyroid cancer cells.65 MiR-335 suppresses cell migration by targeting CPNE1, in NSCLC.31 In small cell lung cancer (SCLC), loss of miR-335 promotes metastatic skeletal lesions through the deregulation of insulin-like growth factor-I receptor (IGF-IR) and receptor activator of nuclear factor-κB ligand (RANKL), key mediators of bone metastases.73 Bcl-w, which is a pro-survival member of the Bcl-2 protein family, is known as a possible target of miR-335 in many cancers. In detail, in NSCLC, the overexpression of miR-335 suppresses the invasion of lung cancer cells by targeting Bcl-w.19 Besides, miR-335 inhibits ovarian cancer cell migration and invasion and leads to depolymerization of F-actin via targeting Bcl-W.21 By the down-regulation of Bcl-W, miR-335 acts as a tumor suppressor to negatively modulate cell proliferation and invasion.44,60 In breast cancer, miR-335 modulated migration and invasion via targeting SIX Homeobox 2 (SIX2).35 Besides, the migration of breast cancer cells is inhibited by miR-335 by targeting the oncoprotein c-Mesenchymal-epithelial transition factor (c-Met), which is a receptor tyrosine kinase.74
Regarding GC, it is found that miR-335 controls invasion by targeting survivin.26 Through down-regulating LDHB, miR-335 suppresses colorectal cancer cell migration and invasion.43 MiR-335 inhibits invasion by modulating the expression of JMJD2C in pancreatic cancer.68 In bladder cancer, miR-335 may exert its migration inhibitory effect by targeting CRKL.46 OCT4 is correlated with the progression of various cancers. MiR-335 inhibits migration and invasion by down-regulating OCT4 in bladder cancer.48 MiR-335 expression in OCT4-positive pancreatic cancer cells is lower compared to that in the negative ones. MiR-335 inhibits migration, EMT, and stem cell properties of pancreatic cancer by reducing OCT4.40
Cell Cycle
The cell cycle is a complex process that consists of a series of events culminating in mitosis and producing two daughter cells. Many molecular pathways and checkpoints can regulate the cell cycle process. Growing studies have verified that miR-335 can regulate cell cycle progression (Figure 2). The Cell cycle is influenced by many factors, such as cyclin D1, cyclin-dependent kinases (CDKs), and CDK inhibitors (CKIs). Cyclin D1 is the key protein of the G1-S phase transition. In melanoma, miR-335 downregulates cyclin D1, Cofilin, and raises Caspase 3, and induces cell cycle arrest by repressing ROCK1.24 MiR-335 decreases the level of cyclin D1 and facilitates cell cycle arrest by inhibiting CUL4B in NSCLC.62 MiR-335 decreases cell division cycle 2 (CDC2) and cell division cycle 25 (CDC25) and induces G0/G1 phase arrest through inhibiting MEF2D.51 CDC2, also known as CDK1, plays a central role in entry and progression through mitosis.75 CDC25 dephosphorylates the threonine 14 (T14) and tyrosine 15 (T15) phosphorylation, thereby activating CDK1.76 Cyclin D1 is significantly up-regulated, however, p21 is significantly down-regulated in miR-335 overexpressing cells in CRC. The P21 is an important CDK inhibitor (CKI) that inhibits CDK1 activity by down-regulating ROCK1, miR-335 acts as a tumor suppressor to modulate cell cycle progression in NSCLC.30 Furthermore, CPNE1 inhibits cell growth by its effects on the cell cycle and is negatively regulated by miR-335 in NSCLC.31
MiR-335 and Cancer Treatments
Chemotherapy and radiation therapy play important roles in cancer treatments, and many pieces of evidence suggest that miR-335 participates in regulating chemotherapy or radiation therapy sensitivity (Figure 3). The decreased expression of histone deacetylase 3 (HDAC3) is possibly responsible for the resistance to anti-cancer drugs. MiR-335 enhances the response to anti-cancer drugs by targeting seven in absentia homolog 2 (SIAH2) and increasing HDAC3.28 The elevated expression of WW domain-binding protein 5 (WBP5) induces multidrug resistance (MDR) under the regulation of miR-335 through the Hippo pathway, in SCLC.77 In osteosarcoma, miR-335 reduces the sphere-forming ability induced by cisplatin by targeting POU class 5 homeobox 1 (POU5F1).78 Poly(ADP-ribose) polymerase 1 (PARP-1) is the main human PARP enzyme broadly involved in the DNA damage response.79 MiR-335 regulates the chemo-radioresistance of SCLC cells by targeting PARP-1.80 Cyclin B1 (CCNB1), a member of the cyclin family, modulates the activity of cycle-independent kinases (CDKs). MiR-335 can enhance gemcitabine sensitivity in RCC by regulating Cyclin B1 (CCNB1).81 Overexpression of miR-335 increases the sensitivity of triple-negative breast cancer (TNBC) cells to paclitaxel, cisplatin and doxorubicin, and improves the effectiveness of chemotherapy.33 MiR-335 improves the cisplatin sensitivity in ovarian cancer, through inhibiting Bcl-w.49 CPNE1 inhibition improves the efficacy of EGFR-tyrosine kinase inhibitors and CPNE1 is a target of miR-335.31 MiR-335 inhibits the C-Met-Akt pathway, and its activation leads to sorafenib resistance in HCC.82 Up-regulated miR-335 enhances the response to irradiation of melanoma cells by targeting ROCK1.24
MiR-335 as an OncomiR
In contrast to the above research works, miR-335 functions as an oncogene to promote cancer progression. In detail, promote cell proliferation and increase cell sensitivity to 5-Fluorouracil (Fu) treatment by down-regulating myocyte enhancer factor 2D (MEF2D) in gallbladder carcinoma.51 MEF2D is a transcription factor that belongs to the MEF2 family. It can regulate cell division, differentiation, and death in muscle, heart, and cancer cells.83 In CRC, miR-335 facilitates cell proliferation and cell cycle progression by targeting RAS p21 protein activator 1 (RASA1), which has been proved to be a cancer suppressor.20
MiR335 and Tumor Microenvironment
The research focusing on the tumor microenvironment of cancer has significantly progressed, indicating that the tumor microenvironment plays a key role in modulating cancer initiation, progression, metastasis, as well as therapeutic responses.84 The tumor microenvironment mainly contains extracellular matrix (ECM) and stromal cells, including blood endothelial cells, lymphatic endothelial cells, cancer-associated fibroblasts (CAFs), tumor-associated macrophages (TAMs), mesenchymal stem cells (MSCs), immune cells, leukocytes, and adipocytes.85,86
CAFs are key players in the tumor microenvironment that contribute to malignant initiation and progression.87 CAFs, which are recognized to be derived from normal fibroblasts (NFs), communicate with tumor cells through numerous growth and inflammation factors.88,89 It is found that up-regulation of miR-335 within fibroblasts of HCC can inhibit neighboring cancer cell proliferation and invasion. This indicates that miR-335 might be shuttled between these fibroblasts and cancer cells.90 Senescent CAFs develop a senescence-associated secretory phenotype (SASP) that is believed to contribute to cancer progression. One study indicates that miR335, overexpressed in the senescent normal fibroblasts and CAFs, can regulate the secretion of SASP factors.91
Tumor cells secrete numerous pro-angiogenic factors to create an abnormal vascular network, resulting in poorly perfused tumors, because of disorganized, immature, and permeable blood vessels.92 Inflammatory responses play pivotal roles in cancer progression, containing initiation, promotion, and metastasis. Cytokines are known as important mediators linking inflammation and tumor.93 Emerging evidence indicates the involvement of miR-335 in angiogenesis and inflammation. In prostate cancer, the upregulated miR-335 significantly reduces the formation of regenerative tubes of DU145 cells and inhibits the expression of inflammatory factors, including IL‑6, IL‑8, and IL‑1β. Early growth response 3 (EGR3) is proposed as a potential target of miR-335 and is negatively modulated by miR-335.47 EGR3, a member of zinc finger transcription factors, is proved to be involved in angiogenesis and inflammation.94,95 These results indicate that miR335 plays an important role in regulating the tumor microenvironment.
The Pathway and Targets of miR335 in Cancer
The Hedgehog Signaling Pathway
The Hedgehog (Hh) signaling pathway plays a key role in embryonic development and aberrant activity of this pathway in adult tissues is thought to be linked to a variety of solid neoplasms.96 HH ligand- secreting cells release several HH isoforms — Sonic hedgehog (Shh), Indian hedgehog, or Desert hedgehog —which can bind to the transmembrane receptors Patched 1 (PTCH1) and PTCH2.97 Thereby the interaction between SHH and PTCH1 suppresses its inhibitory activity against Smoothened (SMO).98 Subsequently, this signaling cascade results in the activation of GLI transcription factors, thereby driving the expression of HH target genes.99 Previous studies have shown that miRNA regulates cancer progression through this signaling pathway.100 One research indicates that miR335 inhibits papillary thyroid carcinoma progression by targeting SHH via suppressing the Hedgehog pathway.65 SHH, one of the ligands of the Hh pathway, has been recognized as involved in tumor growth and metastasis.101
PI3K/AKT/mTOR Signaling Pathway
Emerging studies found that miR-335 participates in most of the oncogenic signaling pathways (Figure 4). The signaling pathway of the phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) is one of the various signaling pathways playing a key role in cell growth and survival.102 The PI3K/AKT/mTOR signaling is activated through binding to the PKT ligand, which includes TGF-β1R, tyrosine-protein kinase Met (c-MET), and epidermal growth factor receptor (EGFR). Another critical component of this pathway is mTOR protein kinase including mTORC1 and mTORC2. It has been reported that the downregulation of miR-335 in lung cancer promotes cell proliferation through activating AKT/mTOR signaling pathway.25 Another study suggests that miR335 may regulate sorafenib sensitivity of HCC cells by targeting C-MET via suppressing the AKT pathway.82 C-MET is one crucial protein tyrosine kinase that plays a critical role in cancer progression.103
Hippo/YAP Signaling Pathway
The hippo/YAP signaling pathway is one of the various signaling pathways, which is identified as a relevant oncogenic signaling pathway in a variety of tumor types.104 Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ), the main effectors of the signaling pathway, are regulated by a series of kinase cascades. YAP acts as a transcription cofactor regulating the expression of genes involved in cell proliferation and migration signals, which contributes to the pro-tumorigenic phenotype.105 Recently, one study suggests miR335 may regulate SCLC MDR through the Hippo pathway. It is found that miR-335 negatively regulates the MDR by targeting WBP5 and the result also indicates that upregulation of WBP5 can inhibit the phosphorylation of MST2 and Yes-associated protein 1 (YAP1), thus inducing the nuclear accumulation of YAP1 in SCLC.77
The Other Targets of miR-335
MiRNAs are involved in various physiological and pathological processes via targeting mRNAs. Apart from the pathways described above, miR-335 regulates the initiation and development of cancer by targeting various targets. According to research, miR335 plays a role as a tumor suppressor via targeting ROCK1 in different tumor types.30,39,56–58 Moreover, BCL-W is also a common target of miR335 in various cancers. Through targeting BCL-W, miR335 regulates tumor growth, metastasis, and drug sensitivity.19,44,60 Meanwhile, in the same type of tumor, miR335 can regulate tumor progression by targeting different genes.20,22,25,26,43,58 According to computer prediction, each miRNA may target about 200 mRNAs.Besides, one protein-encoding gene also can be regulated by various miRNAs, suggesting that miRNAs are vital regulators of mRNAs. Therefore, it is significant to further explore the regulatory network of miR-335 in different human tumors.
Factors Modulating miR-335 in Cancers
LncRNA and CircRNA
The three major members of non-coding RNAs (ncRNAs), miRNAs, long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), play a crucial role in cancer development. Recently, the competing endogenous RNA (ceRNA) mechanism describes lncRNA/circRNA as sponges for miRNAs to indirectly regulate their target mRNAs. Recently, ceRNA mechanism is the main focus among diverse non-coding functions in the tumorigenesis of various cancers. CeRNA model describes a competitive binding between sponge RNAs and miRNA target genes and the modulation of miRNA target gene activity.106 Multiple studies have suggested that lncRNAs or circRNAs, which contain miRNA-binding sites can act as ceRNAs. Notably, this ceRNA mechanism has been recognized in multiple cancers.107,108 Accumulating evidence indicates miR-335 is regulated by a variety of LncRNAs and circRNA in cancers (Table 3).
 |
Table 3 Regulatory Factors Suppress the Expression of miR-335 in Human Cancer |
LncRNA ZNRD1-AS1 is differentially overexpressed and is positively associated with advanced TNM stage and a higher rate of lymph node metastasis. LncRNA ZNRD1-AS1 promotes cell invasion and metastasis by regulating the miR-335–ROCK1 axis in nasopharyngeal carcinoma (NPC).72 Sry-related HMG box 2 (SOX2) modulates LncRNA LINC01510/miR-335/SHH axis to promote thyroid carcinoma progression.65 LncRNA CASC9 expression is elevated in NSCLC and promotes NSCLC progression by regulating the miR-335/S100A14 axis.29 LncRNA-XIST promotes NSCLC progression by targeting miR-335/SOD2/ROS signal pathway induced pyroptotic cell death.64 Platycodin D (PD) modulates LncRNA-XIST/miR-335 axis to suppress bladder cancer progression.109 LncRNA NEAT1 inhibits sorafenib sensitivity of HCC cells by regulating miR-335-cMet.82 Besides, LncRNA NEAT1 causes GC malignancy through down-regulating miR-335 and inducing the expression of ROCK1.58 Besides, high expression of lncRNA MSTO2P is correlated significantly with lymphatic metastasis, distal metastasis, and shorter overall survival rate in GC patients, and lncRNA MSTO2P promotes the proliferation by indirectly regulating miR-335.110 In gastric cancer, lncRNA ZEB1-AS1 negatively regulates miR-335 and plays an important role in promoting GC cell progression by inhibiting miR-335.42 In osteosarcoma, lncRNA DANCR functions as an endogenous sponge for miR-335 and relieves the inhibition of miR-335 on ROCK1.59 Furthermore, by sponging miR-335, LncRNA DANCR elevates ROCK1 and facilitates cervical cancer development.56 LncRNA TUG1 alleviates the inhibitory effect of miR-335 on ROCK1 to promote osteosarcoma progression.71 By sponging miR-335 and regulating CXC chemokine receptor 4 (CXCR4), lncRNA TUG1 promotes tumor progression in CTNNB1-mutated hepatoblastoma.111 LncRNA RP11-436H11.5, functioning as an endogenous sponge for miR-335, upregulates BCL-W by sponging miR-335 and promotes proliferation and invasion in RCC.60 In bladder cancer, lncRNA SLCO4A1-AS1 promotes proliferation and migration by regulating the miR-335/OCT4 axis.48 Besides, LncRNA TINCR functions as a sponge of miR-335 in ovarian cancer, thereby affecting fibroblast growth factor 2 (FGF2) expression to promote cell proliferation, migration, and invasion.112
Increased expression of circ-EIF4G3 is associated with advanced TNM stage and lymphatic metastasis. Circ-EIF4G3 promotes the development of gastric cancer by sponging miR-335.113 Circ_0074027, which is highly expressed in NSCLC, facilitates NSCLC cell progression by modulating miR-335/CUL4B axis.62 Circ_0007255 modulates the progression of breast cancer through miR-335/SIX2 axis.35 In HCC, by repressing miR-335, circ_0009910 regulates ROCK1 and promotes proliferation and metastasis.57 CircZMYM2 modulates the downstream gene JMJD2C by sponging miR-335 and promotes pancreatic cancer cell progression.68 High expression of circ_103973 is related to the worse outcome of cervical cancer patients. Circ_103973 promotes cervical cancer cell proliferation by sponging miR-335, which further regulates PPP6C.70
Other Regulatory Factors
Cadherin 11 (CDH11) belongs to the cadherin superfamily and mediates cell adhesion in a calcium-dependent manner. The high expression of CDH11 is associated with worse overall survival of breast cancer patients. In metastatic breast cancer, the anti-CDH11 antibody inhibits EMT and represses cancer stem cell-like (CSC-like) and metastatic phenotype by up-regulating miR-335.34 A mass of CpG islands exists in the promoter region of miR-335 and Sp1 can negatively modulate the expression of DNA methyltransferases in ovarian cancer cells, indicating that the participation of DNA methylation in the regulation of miR-335. Sp1 facilitates ovarian cancer migration by down-regulating miR-335.114 In HCC, the level of miR-335/MEST methylation is obviously higher in tumors compared with non-tumor tissues. DNA methylation frequently reduces the expression of miR-335 in primary HCC.115
Discussion
Plenty of research studies show that the abnormal expression of miR-335 is detected in a variety of cancers; however, the expression of miR-335 in some cancer types remains controversial. Besides, the effect of miR-335 in various cellular processes and the complex regulatory network for miR-335 is not fully understood. Emerging shreds of evidence indicate that miR-335 can enhance the sensitivity of cancer cells to chemotherapy and radiotherapy. It may be useful as a combination therapy against different cancers.
While the biological function of miR-335 can be further studied through experimental models, its prospect in clinical applications is still unclear. This is not only due to the predictable general obstacles encountered in the application of gene therapy, but also the uncertain adverse effects. Several miRNA-targeted gene therapies have been terminated or withdrawn from clinical trials because of serious adverse effects. For example, the first miRNA replacement therapy clinical trial, which tested the efficacy of miR-34 (MRX34) in cancer, is terminated at Phase I due to severe immune-related side effects.116 In addition, TargomiRs which are minicells-loaded with a miR-16 mimic were investigated in a phase I study. Fortunately, patients with refractory malignant pleural mesothelioma can tolerate TargomiR, despite some dose-limiting toxicity, and adverse effects are observed.117 As for miR-335, more pre-clinical studies, especially, regarding toxicity and safety, should be carried out before applying miR-335-targeted gene therapies for cancers in clinical settings.
Conclusion
In this article, we focus on the scientific achievements of the researchers of miR-335 in human cancer. The dysregulated miR-335 is observed in a variety of cancers, and the expression of miR-335 depends on the type of cancer. Additionally, the level of miR-335 may be used to predict survival in cancer patients. Recent studies reveal that miR-335 is involved in multiple cellular processes, including proliferation, apoptosis, migration, and invasion. Furthermore, miR-335 can affect the sensitivity of cancer cells to chemotherapy and radiotherapy. It has been shown miR-335 may indirectly affect cancer progression upon secretion into the tumor microenvironment. MiR-335 is regulated by various factors such as lncRNAs and microRNAs. Various factors can regulate miR335, such as LncRNAs and circRNAs. In addition to non-coding RNA, DNA methylation and transcription factors contribute to the progression of various cancers as well. In summary, miR-335 has the potential as a biomarker for cancer prognosis or as a treatment target combination with established drugs to enhance drug sensitivity. However, its clinical application has a long way to go.
Acknowledgments
The authors would like to thank colleagues from the department of oncology, the Affiliated Jiangninging Hospital with Nanjing Medical University for their support and feedback.
Funding
This research was funded by the Natural Science Foundation of Jiangsu Province (grant number BK20161110), Key project of Nanjing Science and Technology Plan (grant number ZKX14030), Medical Research Grant of Jiangsu Commission of Health (grant number M2020010), the Science Foundation of Jiangsu Health vocational college (grant number JKC201948), the Science and Technology Development Fund of Nanjing Medical University (grant number NMUB2019235), the Nanjing health science and Technology Development Fund (grant number YKK18201), Science and technology development fund of Nanjing Medical University (grant number 2017NJMUZD091) and the Research and development fund of Kangda College of Nanjing Medical University (grant number KD2017KYJJYB017, KD2020KYJJZD006).
Disclosure
The authors declare that there are no conflicts of interest.
References
1. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi:10.1146/annurev.pathol.4.110807.092222
2. Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi:10.1101/gad.1184404
3. Seitz H. Issues in current microRNA target identification methods. RNA Biol. 2017;14:831–834. doi:10.1080/15476286.2017.1320469
4. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi:10.1053/j.gastro.2007.05.022
5. Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33. doi:10.2174/156652412798376170
6. Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi:10.1074/jbc.M109.031427
7. Tang W, Zhu J, Su S, et al. MiR-27 as a prognostic marker for breast cancer progression and patient survival. PLoS One. 2012;7:e51702. doi:10.1371/journal.pone.0051702
8. Jin C, Wang A, Liu L, Wang G, Li G, Han Z. miR-145-5p inhibits tumor occurrence and metastasis through the NF-κB signaling pathway by targeting TLR4 in malignant melanoma. J Cell Biochem. 2019;120(7):11115–11126. doi:10.1002/jcb.28388
9. Mizuno R, Kawada K, Sakai Y. The molecular basis and therapeutic potential of Let-7 MicroRNAs against colorectal cancer. Can J Gastroenterol Hepatol. 2018;2018:5769591. doi:10.1155/2018/5769591
10. Gulei D, Magdo L, Jurj A, et al. The silent healer: miR-205-5p up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell Death Dis. 2018;9:66. doi:10.1038/s41419-017-0102-8
11. Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi:10.1016/j.bbadis.2011.02.002
12. Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi:10.1016/j.cell.2005.06.036
13. Hydbring P, Wang Y, Fassl A, et al. Cell-cycle-targeting MicroRNAs as therapeutic tools against refractory cancers. Cancer Cell. 2017;31:576–590.e578. doi:10.1016/j.ccell.2017.03.004
14. Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6:8474–8490. doi:10.18632/oncotarget.3523
15. Jovanovic M, Hengartner MO. miRNAs and apoptosis: rNAs to die for. Oncogene. 2006;25:6176–6187. doi:10.1038/sj.onc.1209912
16. Ma L. MicroRNA and metastasis. Adv Cancer Res. 2016;132:165–207.
17. Lou W, Liu J, Gao Y, et al. MicroRNAs in cancer metastasis and angiogenesis. Oncotarget. 2017;8:115787–115802. doi:10.18632/oncotarget.23115
18. Heyn H, Engelmann M, Schreek S, et al. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer. 2011;129:2797–2806. doi:10.1002/ijc.25962
19. Wang H, Li M, Zhang R, et al. Effect of miR-335 upregulation on the apoptosis and invasion of lung cancer cell A549 and H1299. Tumour Biol. 2013;34:3101–3109. doi:10.1007/s13277-013-0878-9
20. Lu Y, Yang H, Yuan L, et al. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol Cell Biochem. 2016;412:235–245. doi:10.1007/s11010-015-2630-9
21. Cao J, Cai J, Huang D, et al. miR-335 represents an invasion suppressor gene in ovarian cancer by targeting Bcl-w. Oncol Rep. 2013;30:701–706. doi:10.3892/or.2013.2482
22. Du W, Tang H, Lei Z, et al. miR-335-5p inhibits TGF-β1-induced epithelial-mesenchymal transition in non-small cell lung cancer via ROCK1. Respir Res. 2019;20:225. doi:10.1186/s12931-019-1184-x
23. Zhang BJ, Gong HY, Zheng F, Liu DJ, Liu HX. Up-regulation of miR-335 predicts a favorable prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:6213–6218.
24. Cheng Y, Shen P. miR-335 acts as a tumor suppressor and enhances ionizing radiation-induced tumor regression by targeting ROCK1. Front Oncol. 2020;10:278. doi:10.3389/fonc.2020.00278
25. Liu J, Bian T, Feng J, et al. miR-335 inhibited cell proliferation of lung cancer cells by target Tra2β. Cancer Sci. 2018;109:289–296. doi:10.1111/cas.13452
26. Yang B, Huang J, Liu H, Guo W, Li G. miR-335 directly, while miR-34a indirectly modulate survivin expression and regulate growth, apoptosis, and invasion of gastric cancer cells. Tumour Biol. 2016;37:1771–1779. doi:10.1007/s13277-015-3951-8
27. Wang Y, Zhao W, Fu Q. miR-335 suppresses migration and invasion by targeting ROCK1 in osteosarcoma cells. Mol Cell Biochem. 2013;384:105–111. doi:10.1007/s11010-013-1786-4
28. Kim Y, Kim H, Park D, Jeoung D. miR-335 targets SIAH2 and confers sensitivity to anti-cancer drugs by increasing the expression of HDAC3. Mol Cells. 2015;38:562–572. doi:10.14348/molcells.2015.0051
29. Zhao W, Chen T, Zhao Y. Upregulated lncRNA CASC9 contributes to progression of non-small cell lung cancer through inhibition of miR-335-3p and activation S100A14 expression. Onco Targets Ther. 2020;13:6027–6036. doi:10.2147/OTT.S249973
30. Tang H, Du W, Jiang Y, Li H, Bo H, Song S. Upregulated expression of ROCK1 promotes cell proliferation by functioning as a target of miR-335-5p in non-small cell lung cancer. J Cell Physiol. 2019. doi:10.1002/jcp.28886
31. Tang H, Zhu J, Du W, et al. CPNE1 is a target of miR-335-5p and plays an important role in the pathogenesis of non-small cell lung cancer. J Exp Clin Cancer Res. 2018;37:131. doi:10.1186/s13046-018-0811-6
32. Ramadan A, Hashim M, Swellam M. Expression of MiR-335 and its target metalloproteinase genes: clinical significance in breast cancer. Arch Physiol Biochem. 2020;1–7. doi:10.1080/13813455.2019.1703004
33. Hao J, Lai M, Liu C. Expression of miR-335 in triple-negative breast cancer and its effect on chemosensitivity. J buon. 2019;24:1526–1531.
34. Chen JH, Huang WC, Bamodu OA, Chang PM, Chao TY, Huang TH. Monospecific antibody targeting of CDH11 inhibits epithelial-to-mesenchymal transition and represses cancer stem cell-like phenotype by up-regulating miR-335 in metastatic breast cancer, in vitro and in vivo. BMC Cancer. 2019;19:634. doi:10.1186/s12885-019-5811-1
35. Jia Q, Ye L, Xu S, et al. Circular RNA 0007255 regulates the progression of breast cancer through miR-335-5p/SIX2 axis. Thorac Cancer. 2020;11:619–630. doi:10.1111/1759-7714.13306
36. Xie Y, Deng H, Wei R, et al. Overexpression of miR-335 inhibits the migration and invasion of osteosarcoma by targeting SNIP1. Int J Biol Macromol. 2019;133:137–147. doi:10.1016/j.ijbiomac.2019.04.016
37. Liu ZF, Liang ZQ, Li L, et al. MiR-335 functions as a tumor suppressor and regulates survivin expression in osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20:1251–1257.
38. Zare A, Ahadi A, Larki P, et al. The clinical significance of miR-335, miR-124, miR-218 and miR-484 downregulation in gastric cancer. Mol Biol Rep. 2018;45:1587–1595. doi:10.1007/s11033-018-4278-5
39. Liu H, Li W, Chen C, Pei Y, Long X. MiR-335 acts as a potential tumor suppressor miRNA via downregulating ROCK1 expression in hepatocellular carcinoma. Tumour Biol. 2015;36:6313–6319. doi:10.1007/s13277-015-3317-2
40. Gao L, Yang Y, Xu H, et al. MiR-335 functions as a tumor suppressor in pancreatic cancer by targeting OCT4. Tumour Biol. 2014;35:8309–8318. doi:10.1007/s13277-014-2092-9
41. Li H, Xie S, Liu M, et al. The clinical significance of downregulation of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tumorigenesis. Int J Oncol. 2014;45:197–208. doi:10.3892/ijo.2014.2415
42. Zhang LL, Zhang LF, Guo XH, Zhang DZ, Yang F, Fan YY. Downregulation of miR-335-5p by long noncoding RNA ZEB1-AS1 in gastric cancer promotes tumor proliferation and invasion. DNA Cell Biol. 2018;37:46–52. doi:10.1089/dna.2017.3926
43. Zhang D, Yang N. MiR-335-5p inhibits cell proliferation, migration and invasion in colorectal cancer through downregulating LDHB. J buon. 2019;24:1128–1136.
44. Wang K, Chen X, Zhan Y, et al. miR-335 inhibits the proliferation and invasion of clear cell renal cell carcinoma cells through direct suppression of BCL-W. Tumour Biol. 2015;36:6875–6882. doi:10.1007/s13277-015-3382-6
45. Cao J, Cai J, Huang D, et al. miR-335 represents an independent prognostic marker in epithelial ovarian cancer. Am J Clin Pathol. 2014;141:437–442. doi:10.1309/AJCPLYTZGB54ISZC
46. Liu XK, Chen D, Li X. MiR-335 suppresses cell proliferation and migration by upregulating CRKL in bladder cancer. Eur Rev Med Pharmacol Sci. 2019;23:2399–2408. doi:10.26355/eurrev_201903_17386
47. Zhang P, Yang X, Wang L, Zhang D, Luo Q, Wang B. Overexpressing miR335 inhibits DU145 cell proliferation by targeting early growth response 3 in prostate cancer. Int J Oncol. 2019;54:1981–1994. doi:10.3892/ijo.2019.4778
48. Yang Y, Wang F, Huang H, Zhang Y, Xie H, Men T. lncRNA SLCO4A1-AS1 promotes growth and invasion of bladder cancer through sponging miR-335-5p to upregulate OCT4. Onco Targets Ther. 2019;12:1351–1358. doi:10.2147/OTT.S191740
49. Liu R, Guo H, Lu S. MiR-335-5p restores cisplatin sensitivity in ovarian cancer cells through targeting BCL2L2. Cancer Med. 2018;7:4598–4609. doi:10.1002/cam4.1682
50. Luo L, Xia L, Zha B, et al. miR-335-5p targeting ICAM-1 inhibits invasion and metastasis of thyroid cancer cells. Biomed Pharmacother. 2018;106:983–990. doi:10.1016/j.biopha.2018.07.046
51. Wang W, Chen LC, Qian JY, Zhang Q. MiR-335 promotes cell proliferation by inhibiting MEF2D and sensitizes cells to 5-Fu treatment in gallbladder carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:9829–9839. doi:10.26355/eurrev_201911_19546
52. Dou XQ, Chen XJ, Zhou Q, Wen MX, Zhang SZ, Zhang SQ. miR-335 modulates Numb alternative splicing via targeting RBM10 in endometrial cancer. Kaohsiung J Med Sci. 2020;36:171–177. doi:10.1002/kjm2.12149
53. Yan Z, Xiong Y, Xu W, et al. Identification of hsa-miR-335 as a prognostic signature in gastric cancer. PLoS One. 2012;7:e40037. doi:10.1371/journal.pone.0040037
54. Wang L, Zhao Y, Xu M, Zhou F, Yan J. Serum miR-1301-3p, miR-335-5p, miR-28-5p, and their target B7-H3 may serve as novel biomarkers for colorectal cancer. J buon. 2019;24:1120–1127.
55. Cui L, Hu Y, Bai B, Zhang S. Serum miR-335 level is associated with the treatment response to trans-arterial chemoembolization and prognosis in patients with hepatocellular carcinoma. Cell Physiol Biochem. 2015;37:276–283. doi:10.1159/000430352
56. Liang H, Zhang C, Guan H, Liu J, Cui Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. 2019;234:7266–7278. doi:10.1002/jcp.27484
57. Li HW, Liu J. Circ_0009910 promotes proliferation and metastasis of hepatocellular carcinoma cells through miR-335-5p/ROCK1 axis. Eur Rev Med Pharmacol Sci. 2020;24:1725–1735. doi:10.26355/eurrev_202002_20349
58. Wang H, Zhang M, Sun G. Long non-coding RNA NEAT1 regulates the proliferation, migration and invasion of gastric cancer cells via targeting miR-335-5p/ROCK1 axis. Pharmazie. 2018;73:150–155. doi:10.1691/ph.2018.7877
59. Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi:10.1186/s12943-018-0837-6
60. Wang K, Jin W, Song Y, Fei X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol Cancer. 2017;16:166. doi:10.1186/s12943-017-0735-3
61. Thirunavukarasou A, Singh P, Govindarajalu G, Bandi V, Baluchamy S. E3 ubiquitin ligase Cullin4B mediated polyubiquitination of p53 for its degradation. Mol Cell Biochem. 2014;390:93–100. doi:10.1007/s11010-014-1960-3
62. Yu C, Ying J, Yu K, Shen W, Jiang M. Circ_0074027 contributes to nonsmall cell lung cancer progression by upregulating CUL4B expression through miR-335-5p. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2020.3579
63. Takeo K, Kawai T, Nishida K, et al. Oxidative stress-induced alternative splicing of transformer 2beta (SFRS10) and CD44 pre-mRNAs in gastric epithelial cells. Am J Physiol Cell Physiol. 2009;297:C330–338. doi:10.1152/ajpcell.00009.2009
64. Liu J, Yao L, Zhang M, Jiang J, Yang M, Wang Y. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging (Albany NY). 2019;11:7830–7846. doi:10.18632/aging.102291
65. Li Q, Wang XJ, Jin JH. SOX2-induced upregulation of lncRNA LINC01510 promotes papillary thyroid carcinoma progression by modulating miR-335/SHH and activating Hedgehog pathway. Biochem Biophys Res Commun. 2019;520:277–283. doi:10.1016/j.bbrc.2019.09.138
66. Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244:164–171. doi:10.1016/j.canlet.2006.03.007
67. Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi:10.1038/nrc2293
68. An Y, Cai H, Zhang Y, et al. circZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem. 2018;51:2224–2236. doi:10.1159/000495868
69. Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068–3074. doi:10.1634/stemcells.2008-0530
70. Zhu Y, Jiang X, Zhang S, Wang L, Zhou Q, Jiang J. Hsa_circ_103973 acts as a sponge of miR-335 to promote cervical cancer progression. Onco Targets Ther. 2020;13:1777–1786. doi:10.2147/OTT.S215736
71. Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi:10.1111/cas.13201
72. Wang Q, Hu X, Du M, et al. ZNRD1-AS1 promotes nasopharyngeal carcinoma cell invasion and metastasis by regulating the miR-335-ROCK1 Axis. Onco Targets Ther. 2020;13:4779–4790. doi:10.2147/OTT.S250028
73. Gong M, Ma J, Guillemette R, et al. miR-335 inhibits small cell lung cancer bone metastases via IGF-IR and RANKL pathways. Mol Cancer Res. 2014;12:101–110. doi:10.1158/1541-7786.MCR-13-0136
74. Gao Y, Zeng F, Wu JY, et al. MiR-335 inhibits migration of breast cancer cells through targeting oncoprotein c-Met. Tumour Biol. 2015;36:2875–2883. doi:10.1007/s13277-014-2917-6
75. Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi:10.1038/nrc2602
76. Krasinska L, Domingo-Sananes MR, Kapuy O, et al. Protein phosphatase 2A controls the order and dynamics of cell-cycle transitions. Mol Cell. 2011;44:437–450. doi:10.1016/j.molcel.2011.10.007
77. Tang R, Lei Y, Hu B, et al. WW domain binding protein 5 induces multidrug resistance of small cell lung cancer under the regulation of miR-335 through the Hippo pathway. Br J Cancer. 2016;115:243–251. doi:10.1038/bjc.2016.186
78. Guo X, Yu L, Zhang Z, Dai G, Gao T, Guo W. miR-335 negatively regulates osteosarcoma stem cell-like properties by targeting POU5F1. Cancer Cell Int. 2017;17:29. doi:10.1186/s12935-017-0398-6
79. Pascal JM. The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst). 2018;71:177–182. doi:10.1016/j.dnarep.2018.08.022
80. Luo Y, Tong L, Meng H, et al. MiR-335 regulates the chemo-radioresistance of small cell lung cancer cells by targeting PARP-1. Gene. 2017;600:9–15. doi:10.1016/j.gene.2016.11.031
81. Yan L, Liu G, Cao H, Zhang H, Shao F. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem Biophys Res Commun. 2019;519:172–178. doi:10.1016/j.bbrc.2019.08.093
82. Chen S, Xia X. Long noncoding RNA NEAT1 suppresses sorafenib sensitivity of hepatocellular carcinoma cells via regulating miR-335-c-Met. J Cell Physiol. 2019.
83. McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi:10.1016/S0968-0004(01)02031-X
84. Tan HY, Wang N, Lam W, Guo W, Feng Y, Cheng YC. Targeting tumour microenvironment by tyrosine kinase inhibitor. Mol Cancer. 2018;17:43. doi:10.1186/s12943-018-0800-6
85. Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi:10.1038/nri3902
86. Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32:303–315. doi:10.1007/s10555-012-9415-3
87. Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev. 2016;30:1002–1019. doi:10.1101/gad.279737.116
88. Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi:10.1016/j.cell.2005.02.034
89. Swartz MA, Iida N, Roberts EW, et al. Tumor microenvironment complexity: emerging roles in cancer therapy. Cancer Res. 2012;72:2473–2480. doi:10.1158/0008-5472.CAN-12-0122
90. Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67:940–954. doi:10.1002/hep.29586
91. Kabir TD, Leigh RJ, Tasena H, et al. A miR-335/COX-2/PTEN axis regulates the secretory phenotype of senescent cancer-associated fibroblasts. Aging (Albany NY). 2016;8:1608–1635. doi:10.18632/aging.100987
92. Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409–426. doi:10.1007/s10456-017-9562-9
93. Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol. 2014;26:54–74.
94. Li S, Miao T, Sebastian M, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37:685–696. doi:10.1016/j.immuni.2012.08.001
95. Liu D, Evans I, Britton G, Zachary I. The zinc-finger transcription factor, early growth response 3, mediates VEGF-induced angiogenesis. Oncogene. 2008;27:2989–2998. doi:10.1038/sj.onc.1210959
96. Steg AD, Katre AA, Bevis KS, et al. Smoothened antagonists reverse taxane resistance in ovarian cancer. Mol Cancer Ther. 2012;11:1587–1597. doi:10.1158/1535-7163.MCT-11-1058
97. Murone M, Rosenthal A, de Sauvage FJ. Hedgehog signal transduction: from flies to vertebrates. Exp Cell Res. 1999;253:25–33. doi:10.1006/excr.1999.4676
98. Nybakken K, Perrimon N. Hedgehog signal transduction: recent findings. Curr Opin Genet Dev. 2002;12:503–511. doi:10.1016/S0959-437X(02)00333-7
99. Pasca Di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi:10.1038/nrc1229
100. Li Y, Zhang D, Chen C, Ruan Z, Li Y, Huang Y. MicroRNA-212 displays tumor-promoting properties in non-small cell lung cancer cells and targets the hedgehog pathway receptor PTCH1. Mol Biol Cell. 2012;23:1423–1434. doi:10.1091/mbc.e11-09-0777
101. Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the Sonic Hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (Basel). 2016;8(2):22. doi:10.3390/cancers8020022
102. Dienstmann R, Rodon J, Serra V, Tabernero J. Picking the point of inhibition: a comparative review of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther. 2014;13:1021–1031. doi:10.1158/1535-7163.MCT-13-0639
103. Jung SN, Kang YE, Lee GH, et al. Brn3a/Pou4f1 functions as a tumor suppressor by targeting c-MET/STAT3 signaling in thyroid cancer. J Clin Endocrinol Metab. 2020;105(9):e3127–e3141. doi:10.1210/clinem/dgaa316
104. Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e310.
105. Liu H, Du S, Lei T, et al. Multifaceted regulation and functions of YAP/TAZ in tumors (Review). Oncol Rep. 2018;40:16–28. doi:10.3892/or.2018.6423
106. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi:10.1016/j.cell.2011.07.014
107. Zhao L, Han T, Li Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB j. 2017;31:893–903. doi:10.1096/fj.201600994R
108. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi:10.1186/s12943-018-0827-8
109. Chen D, Chen T, Guo Y, Wang C, Dong L, Lu C. Platycodin D (PD) regulates LncRNA-XIST/miR-335 axis to slow down bladder cancer progression in vitro and in vivo. Exp Cell Res. 2020;396:112281. doi:10.1016/j.yexcr.2020.112281
110. Li H, Zhu H, Zhou Y, et al. Long non-coding RNA MSTO2P promotes the proliferation and colony formation in gastric cancer by indirectly regulating miR-335 expression. Tumour Biol. 2017;39:1010428317705506.
111. Xie F, Zhang L, Yao Q, et al. TUG1 promoted tumor progression by sponging miR-335-5p and regulating CXCR4-mediated infiltration of pro-tumor immunocytes in CTNNB1-mutated hepatoblastoma. Onco Targets Ther. 2020;13:3105–3115. doi:10.2147/OTT.S234819
112. Li R, Wang Y, Xu Y, He X, Li Y. Silencing the long noncoding RNA, TINCR, a molecular sponge of miR335, inhibits the malignant phenotype of epithelial ovarian cancer via FGF2 suppression. Int J Oncol. 2019;55:1110–1124. doi:10.3892/ijo.2019.4875
113. Wang Q, Wang T, Hu Y, et al. Circ-EIF4G3 promotes the development of gastric cancer by sponging miR-335. Pathol Res Pract. 2019;215:152507. doi:10.1016/j.prp.2019.152507
114. Wang S, Li Y, Sun S, Cai J, Cao J. Sp1 promotes ovarian cancer cell migration through repressing miR-335 expression. Biochem Biophys Res Commun. 2020;524:211–216. doi:10.1016/j.bbrc.2020.01.063
115. Dohi O, Yasui K, Gen Y, et al. Epigenetic silencing of miR-335 and its host gene MEST in hepatocellular carcinoma. Int J Oncol. 2013;42:411–418. doi:10.3892/ijo.2012.1724
116. Slabakova E, Culig Z, Remsik J, Soucek K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017;8:e3100. doi:10.1038/cddis.2017.495
117. van Zandwijk N, Pavlakis N, Kao SC, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, Phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi:10.1016/S1470-2045(17)30621-6
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.