Back to Journals » Cancer Management and Research » Volume 11
FRMPD1 activates the Hippo pathway via interaction with WWC3 to suppress the proliferation and invasiveness of lung cancer cells
Authors Rong X, Han Q , Lin X, Kremerskothen J, Wang E
Received 14 November 2018
Accepted for publication 17 March 2019
Published 18 April 2019 Volume 2019:11 Pages 3395—3410
DOI https://doi.org/10.2147/CMAR.S194512
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Xuezhu Rong,1 Qiang Han,1 Xuyong Lin,1 Joachim Kremerskothen,2 Enhua Wang1
1Department of Pathology, College of Basic Medical Sciences and First Affiliated Hospital, China Medical University, Shenyang, People’s Republic of China; 2Internal Medicine D, Department of Nephrology, Hypertension and Rheumatology, University Hospital Muenster, Muenster, Germany
Purpose: The expression of FERM-domain-containing protein-1 (FRMPD1)/FERM and PDZ domain-containing protein-2 (FRMD2) in malignant tumors, including lung cancer, and its underlying molecular mechanism have not been reported yet.
Materials and methods: Immunohistochemistry was performed to analyze the expression of FRMPD1 in lung cancer tissues, and statistical analysis was applied to analyze the relationship between FRMPD1 expression and clinicopathological factors. The biological effects of FRMPD1 on lung cancer cell proliferation and invasion were determined by functional experiments both in vivo and in vitro. Immunoblotting, RT-qPCR, dual-luciferase assay, and immunofluorescence were performed to demonstrate whether FRMPD1 stimulates Hippo signaling. Co-immunoprecipitation assays were used to clarify the underlying role of FRMPD1 in Hippo pathway activation via interaction with WW and C2 domain containing protein-3 (WWC3).
Results: We found that FRMPD1 expression in lung cancer specimens was lower than that in normal bronchial epithelium and normal submucosal glands. FRMPD1 expression had a negative correlation with age, Tumor-Node-Metastasis (TNM) stage, lymph node metastasis, as well as poor prognosis. Moreover, ectopic expression of FRMPD1 significantly inhibited the proliferation and invasion of lung cancer cells, and inhibition of FRMPD1 expression led to opposite effects. Mechanistically, we found that FRMPD1 interacted with the C-terminal PDZ binding motif of WWC3 via its PSD95/DLG/ZO1 (PDZ) domain and promoted the phosphorylation of large tumor suppressor-1 (LATS1), thus inhibiting the nuclear translocation of yes-associated protein (YAP).
Conclusion: FRMPD1 could activate the Hippo pathway and ultimately inhibit the malignant behavior of lung cancer cells through its interaction with WWC3. This work will provide an important experimental basis for the discovery of novel biomarkers of lung cancer and the development of targeted drugs.
Keywords: FRMPD1, Hippo pathway, LATS1, NSCLC, WWC3
Introduction
The Hippo pathway, first discovered in Drosophila melanogaster, is highly conserved across species.1 This pathway regulates not only organ size but also cell proliferation, differentiation, and apoptosis.2,3 In recent years, increasing attention has been focused on the relationship between the Hippo pathway and the proliferation, invasion, and metastasis of tumor cells.4 The Hippo pathway includes upstream molecules (Fat, NF2, WWCs, AMOT, etc.), central kinase complex (MST1/2-WW45-LATS1/2-MOB), downstream effector molecules, yes-associated-protein (YAP), and the target genes of the Hippo pathway, including connective tissue growth factor (CTGF), CyclinE, and DIAP. When the Hippo pathway is activated, the MST1/2-LATS1/2 kinase complex undergoes cascade phosphorylation, which promotes the phosphorylation of YAP (p-YAP). The p-YAP remains in the cytoplasm, where it binds to the 14-3-3 protein and degrades it through the ubiquitin-proteasome pathway. When the Hippo pathway is inhibited, the non-phosphorylated YAP binds to the transcriptional factor TEADs (TEA-domain transcription factors) and promotes the transcriptional activity of its target genes, including CTGF, CyclinE, and CYR61, thereby enhancing the proliferation and invasion of tumor cells.5–7
WWC3 is one of the three members of the WWC protein family (WWC1, WWC2, and WWC3); all the members of this family share similar protein structures, including an N-terminal dual WW domain (which binds to the PPxY motif), C2 domain in the central part, and a C-terminal PDZ domain binding motif.8,9 Our previous study confirmed that WWC3 can regulate the activity of both the Wnt and Hippo pathways by interacting with DVLs and LATS1 to inhibit the proliferation, invasion, and metastasis of lung cancer cells.10 FRMPD1, also called FRMD2, belongs to the FERM family (FERM-domain-containing proteins).11,12 FRMPD1 is located on the 9p13.2 position of chromosome 9. The full length of this gene is 4731 bp, and the molecular weight of the FRMPD1 protein is 169 kDa (GenBank® accession number NM_014907.2). The main domains include the FERM domain (Leu177-Phe401) and the PDZ domain (Gln67-Thr135).13 Current research available on the FERM family is mainly focused on the FRMD6 protein. For example, in Drosophila melanogaster, FRMD6 binds to the C-terminal of Merlin, an upstream protein of the Hippo pathway, via its FERM domain, thereby promoting the Hippo pathway, retaining Yki in the cytoplasm, and in turn, regulating organ sizes in Drosophila melanogaster.14–16 In addition, overexpression of FRMD6 in the YAP-induced MCF-10A cell line of normal ductal epithelial cells of human breast can activate the Hippo pathway, inhibit the epithelial-mesenchymal transition induced by the nuclear translocation of YAP, and ultimately, inhibit the malignant progression of cancer cells.17 To date, the expression of FRMPD1 in human malignant tumors and the association between FRMPD1 and the Hippo pathway have not been reported.
Although there have been few studies on the role of FRMPD1 in human malignancies, analysis of FRMPD1 and WWC3 domains has shown a possible interaction between the PDZ domain in FRMPD1 and the PDZ domain-binding motif in WWC3.10 We speculated that because WWC3 plays an important role in the Hippo pathway, FRMPD1 may regulate this pathway by binding to WWC3, thereby affecting the malignant phenotype of lung cancer cells. In this study, we first examined the expression of FRMPD1 in lung cancer cell lines and resected specimens. Then, we detected the effect of bidirectional regulation of FRMPD1 on the proliferation and invasion of lung cancer cells. Finally, we generated mutants of FRMPD1 and WWC3 to detect whether FRMPD1 interacts with WWC3 through its corresponding domains. This study revealed the specific molecular mechanism underlying the regulation of the Hippo pathway via FRMPD1.
Material and methods
Specimen collection
We collected 127 tumor specimens, including non-small cell lung cancer (NSCLC) tissues and paired non-tumor tissues (20 patients; >5 cm distal to the primary tumor’s edge) from patients (average age: 60 years) who underwent surgery at the First Affiliated Hospital of China Medical University from 2005 to 2014. Written informed consent was obtained from all patients, and all procedures were approved by the Institute Research Ethics Committee (No. 2015 [LS] 023, China Medical University). All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. All tumor specimens were obtained during surgical resection, and all patients were chemotherapy- and radiotherapy-naive prior to resection. A total of 55 and 72 patients presented with adenocarcinoma and squamous cell carcinoma, respectively, per the 2015 classification criteria of the World Health Organization for lung cancer.18 According to the 2010 International Union of cancer TNM staging standards,19 51 tumors were classified as phase I and II, and 76 tumors were classified as phase III.
Western blot analysis (WB) and immunoprecipitation (IP)
Assays were performed as described previously.20 WB analysis and IP were carried out with the following antibodies: FRMPD1 (Sigma, St. Louis, MO, USA, #SAB1407148, 1:500/WB); MYC-tag (Cell Signaling Technology Inc., Danvers, MA, USA, #2276, 1 µg/IP, 1:1,000/WB); GFP (Clontech, CA, USA, #JL-8, 1 μg/IP, 1:3,000/WB); GAPDH (Cell Signaling Technology, #5174, 1:1000/WB); MST1 (Cell Signaling Technology, #3682, 1:1,000/WB); p-MST1 (Cell Signaling Technology, #49332, 1:500/WB); LATS1 (Cell Signaling Technology, #9153, 1:1,000/WB); p-LATS1 (Cell Signaling Technology, #8654, 1:500/WB); YAP (Cell Signaling Technology, #4912, 1:1,000/WB); p-YAP (Cell Signaling Technology, #4911, 1:1,000/WB); LaminB1 (Abcam, Cambridge, MA, USA, #ab16048, 1:1,000/IB); Tubulin (Abcam, #ab52866, 1:1,000/IB). The peroxidase-coupled secondary antibodies were purchased from Santa Cruz Biotechnology Inc. (SantaCruz, CA, USA). Target proteins on PVDF membrane (EMD Millipore, Billerica, MA, USA) were visualized using an ECL kit (Thermo Fisher Scientific, Waltham, MA, USA) and images were obtained using the Bio-Rad Imaging System (Bio-Rad, Hercules, CA, USA).
Immunohistochemistry (IHC)
Assays were performed as described previously.21 Two investigators, who were blinded to the clinical data, examined all the of tumor slides. Five random fields of view were examined per slide, and 100 cells were observed per view at 400× magnification. Briefly, tissue sections were incubated with the FRMPD1 rabbit polyclonal antibody (#HPA042934, 1:200; Sigma, St. Louis, MO, USA). The intensity of the FRMPD1 staining was scored as follows: 0 (no staining), 1 (weak), 2 (moderate), or 3 (high). Percentage scores were assigned as follows: 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The scores of each tumor sample were multiplied to give a final score of 0–12. Tumor samples with scores ≥4 were considered to have strong positive FRMPD1 expression, tumors with scores between 1 and 4 were considered to have weak expression, and tumors with scores of 0 were considered to have negative expression. Phosphate-buffered saline (PBS, Maixin, Fuzhou, China) and goat serum (Maixin, Fuzhou, China) were used as negative controls.
Cell culture
The human immortalized bronchial epithelial (HBE) cell line was obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The LK2 cell line was a gift from H. Kijima (Department of Pathology and Bioscience, Hirosaki University Graduate School of Medicine, Japan); this cell line was approved by the Institute Research Ethics Committee of the China Medical University. All other cell lines were obtained from the Shanghai Cell Bank (Shanghai, China) and cultured in 10% Fetal Bovine Serum (Gibco, New York, USA) of Roswell Park Memorial Institute (RPMI)-1640 medium, 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich) at 37°C and 5% CO2 with high humidity. All cell lines were authenticated by short tandem repeat DNA profiling.
Plasmid construction and transfection
The pEGFP-C2 empty vector, pEGFP-C2-WWC3 plasmid, and pEGFP-C2-WWC3-ΔADDV plasmid were gifts from Professor Joachim Kremerskothen (Muenster University, Germany). The CMV region was the promoter of the pEGFP-C2 vector, and it was isolated and purified by using Kanamycin (50 µg/mL, TransGen Biotech, Beijing, China). The Myc-DDK-tagged-FRMPD1 plasmid was purchased from Origene (Rockville, MD, USA). Both Myc-DDK-tagged-FRMPD1-ΔPDZ and siRNA-FRMPD1 were constructed by Ribobio (Guangzhou, China). Prior to transfection, cells were cultured in medium-deprived serum for 3 h, and Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used for plasmid transfection per the manufacturer’s instructions. Stable transfection was screened by G418 (#A1720, Sigma, St. Louis, MO, USA).
Nuclear and cytosolic fractionation
Cultured cells were trypsinized and washed with cold PBS, and the cell pellets were resuspended in ice-cold lysis buffer (210 mM mannitol, 70 mM sucrose, 5 mM Tris, pH 7.5, 1 mM EDTA supplemented with protease inhibitors). After being incubated on ice for 15 min, the cells were homogenized. The nuclei were separated by centrifugation (10 min, 12,000 round/min, 4 °C), and the supernatant containing the cytosolic fraction was boiled in sample buffer. The pellet containing the nuclei was washed with PBS and then resuspended in RIPA buffer (Santa Cruz Biotechnology, CA, USA) for 5 min on ice. The pellet was then centrifuged again, and the supernatant (nuclear fraction) was boiled in sample buffer. Finally, the cytosolic and nuclear fractions were detected by WB.
Immunofluorescence staining
Assays were performed as described previously.21 Briefly, plasmids and siRNA were transfected into cells. After 48 h, cells were fixed, permeabilized, and incubated with primary antibodies and fluorescein isothiocyanate-conjugated (FITC) or tetramethyl rhodamine isothiocyanate-conjugated (TRITC) secondary antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Slides were observed under an inverted Nikon TE300 microscope (Melville, NY, USA). Five random fields of view were examined per slide and the final images were obtained and analyzed using confocal microscopy with FLUOVIEW viewer software.
Colony formation, matrigel invasion, and MTT assays
Colony formation
Forty-eight hours after transfection with plasmids or siRNA, cells were seeded in three 6-cm cell culture dishes (1,000 cells/dish) and incubated for 12 days in 10% FBS of RPMI-1640 medium. The plates were then washed with PBS and stained with Giemsa (Beyotime Biotechnology, Shanghai, China) before counting the number of colonies consisting of >50 cells.
Matrigel invasion
A cell invasion assay was performed using a 24-well Transwell chamber with an 8 μm pore size (Thermo Fisher Scientific, Waltham, MA, USA), and the inserts were coated with 20 μL Matrigel (1:3 dilution, BD Bioscience, New Jersey, USA). LK2 cells transfected with the FRMPD1 plasmid or FRMPD1 shRNA for 48 h were trypsinized, transferred to the upper Matrigel chamber in 100 μL of serum-free medium containing 5×105 cells, and incubated for 16 h. Medium supplemented with 10% FBS was added to the lower chamber as the chemoattractant. The number of invaded cells was counted in 10 randomly selected high-power fields under a microscope (Nikon TE300, Melville, NY, USA).
MTT assays
Forty-eight hours after transfection, cells were plated in 96-well plates in media containing 10% FBS at about 2,000 cells/well, and cell viability was determined using the MTT assay. Briefly, 20 μL of 5 mg/mL MTT solution (Sigma, St. Louis, MO, USA) was added to each well and incubated for 4 h at 37 °C. Then, the medium was removed from each well and the resultant MTT formazan was solubilized in 150 μL of DMSO (Sigma, St. Louis, MO, USA). The results were quantified spectrophotometrically (Bio-Rad Technology, CA, USA) using a test wavelength of 490 nm.
RNA extraction and real-time RT-PCR (RT-qPCR)
Cells were transfected with the indicated plasmid or siRNA. After 48 h, RNA was extracted and RT-qPCR assays were performed as described previously.20 The relative transcript levels of genes were normalized to GAPDH mRNA levels. Primers were synthesized (Sangon Biotech, Shanghai, China) using the sequences listed below:
FRMPD1: 5′-TGCGACACACAGTAAAGATAGAC-3′ (forward)
5′-GAGAATATCGACTGCTCGTTCC-3′ (reverse)
CYR61: 5′-CTCGCCTTAGTCGTCACCC-3′ (forward)
5′-CGCCGAAGTTGCATTCCAG-3′ (reverse)
CTGF: 5′-AACTGCAACCTCTCGCACTG-3′ (forward)
5′-GCTCGGGCTCCTTGTAATTCT-3′ (reverse)
GAPDH: 5′-GGAGCGAGATCCCTCCAAAAT-3′ (forward)
5′-GGCTGTTGTCATACTTCTCATGG-3′ (reverse)
Dual-luciferase assay
YAP/TEAD transcriptional activity was measured using a luciferase assay based on the pGL3b_8xGTIIC-luciferase plasmid obtained from Addgene (plasmid #34615, Cambridge, MA, USA). Cells were transfected to express the indicated proteins and Renilla luciferase was used as a control for signal normalization. The dual luciferase assays were performed according to the manufacturer’s protocol (Progema, Madison, Wisconsin, USA). Three independent transfections were carried out for each experiment. Data were normalized to the empty vector control and presented as average ± SD.
Transplantation of tumor cells into nude mice
The nude mice used in this study were treated following the experimental animal ethics guidelines issued by the China Medical University. The study was approved by the Institutional Animal Research Committee of China Medical University. Four-week-old female BALB/c nude mice were purchased from Charles River (Beijing, China), and the axilla or tail vein of each mouse was subcutaneously or intravenously inoculated with 5×106 or 2×106 tumor cells, respectively, in 0.2 mL of sterile PBS. Six weeks after inoculation, mice were euthanized and autopsied to examine tumor growth and dissemination. A portion of tissue from the tumor and each organ was fixed in 4% formaldehyde (Sigma) and embedded in paraffin. Serial 4-μm-thick sections were prepared and stained with hematoxylin and eosin (H&E), and the stained sections were examined under a microscope (Nikon TE300, Melville, NY, USA).
Statistical analysis
The statistical software SPSS 22.0 (statistical program for social sciences 22.0, Chicago, IL, USA) was used for all analyses. The chi-square test was used to assess correlations between FRMPD1 expression and clinicopathological factors. The Cox regression model was used to test the prognostic value. All clinicopathological parameters were included in the Cox regression model and tested by univariate analysis using the enter method and by multivariate analysis using the forward stepwise logistic regression method. Differences between the groups were tested with a Student’s t-test(Paired t-test). A P-value of <0.05 was considered to be significant.
Results
Low expression of FRMPD1 in lung cancer is associated with poor prognosis
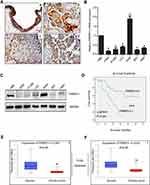
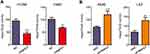
To investigate the expression pattern of FRMPD1 in lung cancer tissues, we collected 127 NSCLC specimens. Immunohistochemical staining showed that FRMPD1 was localized in the cytoplasm of lung cancer cells and was strongly expressed in normal bronchial epithelium and normal submucosal glands (15/20, 75%); it was negatively or weakly expressed in squamous cell carcinoma and adenocarcinoma (61.4%, 78/127. Positive expression was 38.6%, 49/127. Figure 1A). The low expression of FRMPD1 was associated with age (P=0.040), lymph node metastasis (P<0.001), and advanced TNM stage (P<0.001) in lung cancer patients (Table 1). WB, RT-qPCR, and confocal microscopy were employed to detect the protein levels, mRNA levels, and location of FRMPD1, respectively, in six lung cancer cell lines (A549, H1299, SPC, H460: lung adenocarcinoma, LK2: lung squamous cell carcinoma, H661: large cell lung cancer) and the HBE cell line. Results showed that the expression of FRMPD1 in five lung cancer cell lines (5/6) was significantly lower than that in the HBE cell line, and that FRMPD1 was localized in the cytoplasm at the same time (Figure 1B and C; Figure S1A). In addition, Kaplan-Meier survival analysis showed that patients with FRMPD1 positive expression (49.540 months ±3.044) survived significantly longer than those (28.178 months ±1.263) with negative expression (P<0.001, Figure 1D). Cox regression analysis indicated that low FRMPD1 expression was an independent risk factor for poor prognosis of NSCLC patients (P<0.05, Table 2). Finally, the TCGA database (
 | Table 1 Analysis of clinicopathological factors |
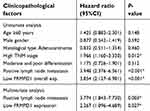 | Table 2 Cox univariate and multivariate risk factor analysis |
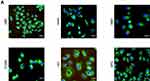 | Figure S1 FRMPD1 is localized in the cytoplasm. Confocal microscope showed that FRMPD1 was localized in the cytoplasm in six cell lines (A), Bar A: 50 µm, magnification: 200×. |
FRMPD1 can inhibit proliferation and invasion of lung cancer cells
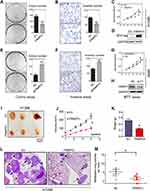
To further explore the effects of FRMPD1 on the malignant phenotype of lung cancer cells, we first transfected FRMPD1 into H1299 and H460 cell lines with low expression of FRMPD1, and then carried out colony formation, MTT viability, and Transwell invasion assays. The results confirmed that ectopic expression of FRMPD1 inhibited the proliferation (H1299: Control group vs FRMPD1 group, 151.12±5.774 vs 51.34±11.55, P=0.0015; H460: Control group vs FRMPD1 group, 212.7±4.055 vs 105.7±3.480, P<0.001) and invasion (H1299: Control group vs FRMPD1 group, 101.7±2.028 vs 53.00±7.371, P=0.0031; H460: Control group vs FRMPD1 group, 158.0±16.77 vs 89.00±2.082, P=0.0039) of tumor cells (Figure 2A–D; Figure S2A–D). In contrast, when FRMPD1 was knocked down in A549 and LK2 cell lines with relatively high expression of FRMPD1, the proliferation (A549: Control group vs siFRMPD1 group, 46±7.810 vs 108.7±7.311, P=0.0042; LK2: Control group vs siFRMPD1 group, 100.3±2.603 vs 202.0±2.517, P<0.001) and invasion (A549: Control group vs siFRMPD1 group, 169.7±6.692 vs 240.0±6.658, P=0.0017; LK2: Control group vs siFRMPD1 group, 108.0±4.359 vs 305.3±5.840, P<0.001) of tumor cells were enhanced (Figure 2E–H; Figure S2E–H). To assess the effect of FRMPD1 on the proliferation and invasion in vivo, the H1299 cell line stably expressing FRMPD1 (selected by G418) was subcutaneously injected or intravenously injected (via the tail vein) into nude mice. Although the tumor formation rate was similar (3/3, 100%), the volume and weight of subcutaneously injected tumors were reduced in the FRMPD1 group as compared with the control group (control group vs FRMPD1 group, weight: 0.5257 g ±0.052 vs 0.2543 g ±0.03, P<0.05. Figure 2I–K). After intravenous injection, as compared with the control group, FRMPD1 overexpression reduced intrapulmonary metastasis formation (control group vs FRMPD1 group: 6.1±1.2 vs 2.5±0.7, n=7, P<0.05. Figure 2L and M). The above results indicated that FRMPD1 inhibits the proliferation and invasion of tumors. Because FRMD6/Willin, which belong to the same family, can activate the Hippo pathway by binding to Merlin, it can be speculated that FRMPD1 may also inhibit tumor proliferation and invasion by activating the Hippo pathway.
FRMPD1 inhibits nuclear translocation of YAP by activating the Hippo pathway
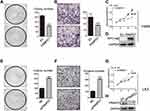
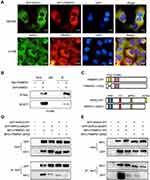
According to previous reports,17 FRMD6/Willin can activate the Hippo pathway by binding to Merlin, an upstream molecule of the Hippo pathway. Therefore, in the interest of exploring the molecular mechanism through which FRMPD1 affects malignant lung cancer phenotypes, we focused on the effects of FRMPD1 on the Hippo pathway. We transfected FRMPD1 plasmids into H1299 and H460 cell lines and found the luciferase activity of YAP-TEAD to be significantly downregulated (Figure S3A). At the same time, WB showed that the phosphorylation of LATS1 and YAP was upregulated, whereas expression levels of CTGF and CyclinE in the nucleus were downregulated (Figure 3A). In contrast, the phosphorylation of LATS1 and YAP was decreased; furthermore, the luciferase activity of YAP-TEAD and the expression levels of CTGF and CyclinE in the nucleus of the Hippo pathway were enhanced after silencing of FRMPD1 in A549 and LK2 (Figure 3; Figure S3). In H1299 cell lines, transfection of FRMPD1 inhibited the nuclear translocation of YAP, as demonstrated by nucleus-cytoplasm isolation and immunofluorescence (Figure 3C and D). In A549 cells, silencing of FRMPD1 promoted the nuclear translocation of YAP (Figure 3E and F). Therefore, we concluded that FRMPD1 stimulates the phosphorylation of the LATS1-YAP cascade to retain YAP in the cytoplasm, thus inhibiting the expression of CTFG and CyclinE and activating the Hippo pathway.
FRMPD1 activates the Hippo pathway through binding of its PDZ domain and the ADDV domain of WWC3
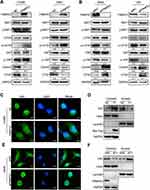
By analyzing the molecular structure of WWC3 (upstream protein of the Hippo pathway) and FRMPD1, we speculated that the PDZ domain of FRMPD1 might interact with the PDZ binding motif (ADDV domain) of WWC3. To confirm this hypothesis, we performed a confocal microscopy assay and revealed that the two proteins had exogenous and endogenous co-localization in the cytoplasm of HEK293 and H1299 cell lines (Figure 4A). Next, we co-transfected GFP-WWC3 and MYC-FRMPD1 plasmids into the H1299 cell line; the co-immunoprecipitation assay showed that MYC-FRMPD1 interacts with GFP-WWC3 (Figure 4B). To determine the binding sites involved in the interactions between FRMPD1 and WWC3, we next constructed a series of FRMPD1 (MYC-FRMPD1-ΔPDZ) and WWC3 (GFP-WWC3-ΔADDV) mutant plasmids (Figure 4C). Co-immunoprecipitation results showed that FRMPD1-ΔADDV could not interact with WWC3; on the other hand, WWC3 could not interact with FRMPD1 when it lacked a carbon-terminal ADDV binding motif (Figure 4D and E). Based on the above results, we concluded that FRMPD1 interacts with the carbon-terminal ADDV binding motif of WWC3 via its own PDZ domain.
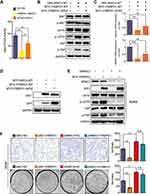
FRMPD1 inhibits the malignant phenotype of lung cancer by activating the Hippo pathway via interaction with WWC3
Our previous studies have demonstrated that WWC3 can activate the Hippo pathway via LATS1 in lung cancer cell lines and that it serves as an upstream activator of the Hippo pathway.10 However, how does the interaction between FRMPD1 and WWC3 affect the activity of the Hippo pathway to inhibit the proliferation and invasion of lung cancer cells? To answer this question, first we co-transfected wild-type WWC3 and FRMPD1 and their mutants into a H1299 cell line exhibiting low expression of WWC3, as demonstrated in our previous work.10 Through a luciferase reporter and WB assays, we found that wild-type FRMPD1 interacted with WWC3 to promote the activity of the Hippo pathway (through downregulation of YAP-TEAD transcriptional activity) and to upregulate the phosphorylation levels of LATS1 and YAP; however, the absence of a PDZ domain in FRMPD1, which hindered the interaction with WWC3, abrogated these effects (Figure 5A and B). In addition, RT-qPCR showed that co-transfection of MYC-FRMPD1-WT and GFP-WWC3-WT significantly downregulated mRNA expression levels of the YAP downstream genes, CTGF and CYR61, whereas co-transfection of FRMPD1 mutants and wild-type WWC3 did not show any such effects (Figure 5C and D). Finally, we transfected the FRMPD1 plasmid into A549 cells with relatively high WWC3 expression and knocked down WWC3 using siRNA.10 The results showed that FRMPD1 could not significantly increase the phosphorylation levels of LATS1 and YAP when WWC3 was silenced (Figure 5E); furthermore, the inhibitory effect on lung cancer cell proliferation and invasion was also significantly weakened (n.s.: Transwell: p=0.0512; Colony formation: p=0.0598 Figure 5F). Consequently, we concluded that FRMPD1 inhibited the malignant phenotype of lung cancer cells by interacting with WWC3 and activating the Hippo pathway.
Discussion
In recent years, the incidence and mortality rate of lung cancer have been the highest among all kinds of malignant tumors,22 and about 80% of deaths are reportedly due to invasion and metastasis.23 Therefore, revealing the molecular mechanism underlying the invasion and metastasis of lung cancer and searching for new biomarkers of biological behavior of lung cancer cells would not only benefit the prognostic evaluation of lung cancer but also provide an experimental basis for the development of effective targeted therapeutic drugs, which have important theoretical and practical significance.
The expression of FRMPD1 in malignant tumors, especially in lung cancer, and its possible biological effects have not yet been reported. In this study, we first found that FRMPD1 expression was decreased in lung cancer resected specimens through immunohistochemistry, and that FRMPD1 expression was negatively correlated with a patient’s age, high TNM stage, and lymph node metastasis. Kaplan-Meier survival analysis showed that FRMPD1-positive patients survived significantly longer than FRMPD1-negative patients, suggesting that FRMPD1 could be recognized as a prognostic factor for lung cancer. However, whether it is also suitable for other types of cancer needs further study. The TCGA database also showed that FRMPD1 was weakly expressed in lung adenocarcinomas. Although the expression levels of FRMPD1 in lung squamous cell carcinoma were comparable to those in normal lung tissues, its expression tended to decrease, which may be related to the selection of samples. Overexpression of FRMPD1 in lung cancer cell lines significantly inhibited the proliferation and invasion of lung cancer cells. These data suggested that FRMPD1 might play a role as a tumor suppressor gene in the development of lung cancer. However, the reason for the low expression of FRMPD1 in non-small cell lung cancer is still unclear. The possibility of methylation or mutation of the FRMPD1 gene or metabolic enhancement in lung cancer cells needs to be explored and verified in subsequent experiments.
To explore the molecular mechanism by which FRMPD1 affects the malignant phenotype of lung cancer, we explored the effects of FRMPD1 on the Hippo pathway and the phosphorylation levels of the key proteins LATS1 and YAP. As observed by RT-qPCR, luciferase reporter, and immunoblotting assays, FRMPD1 significantly enhanced the phosphorylation of LATS1 and YAP, and inhibited the nuclear translocation of YAP and the transcriptional activity of the target gene. Moreover, FRMPD1 was confirmed to be a positive regulator of the Hippo pathway. Protein-protein interactions play a vital role in biological functions. The protein WWC3 contains a PDZ binding motif (which can bind to the PDZ domain), and FRMPD1 contains a PDZ domain, observations worth exploring. Using co-immunoprecipitation and confocal microscopy, we found that FRMPD1 interacts with WWC3 via its N-terminal PDZ domain (possibly direct binding, not yet demonstrated), and that FRMPD1 and WWC3 co-localize in the cytoplasm of lung cancer cells. We found that there were no significant changes in the activity of the Hippo pathway, phosphorylation index, and expression levels of target genes after transfection of FRMPD1 and silencing of WWC3, and that the transfection of FRMPD1-ΔPDZ (which could not bind to WWC3) did not stimulate the Hippo pathway.
Several studies have reported that the Hippo pathway is involved in immunotherapy to malignant tumors, such as lung cancer and melanoma. For example, LATS1/2 deletion unmasks a malignant cell’s immunogenic potential and retains tumor growth due to the induction of anti-tumor immune responses.24 TAZ can upregulate the expression of PD-L1 and increase the activity of the PD-L1 promoter.25 At the same time, in human NSCLC, YAP plays an important role in regulating the tumor immune checkpoint in the PD-L1/PD-1 pathway.26 All these conclusions suggest that the Hippo pathway may have an important effect in cancer immunotherapy. Although there are no reports on the effect of FRMPD1 in carcinoma, our study proved that FRMPD1 may inhibit an advanced TNM stage, lymph node metastasis, and poor prognosis of patients in lung cancer by upregulating the phosphorylation of LATS1 and inhibiting the nuclear translocation of YAP; this activates the Hippo pathway and ultimately inhibits the proliferation and invasion of lung cancer cells. Therefore, whether FRMPD1 can be used as a target of immunotherapy warrants further investigation. In conclusion, our results provide important insights in the search for biomarkers for lung cancer, the development of targeted drugs, and elucidation of the mechanism leading to drug resistance in lung cancer.
Abbreviation list
FRMPD1, FERM-domain-containing protein-1; WWC3, WW and C2 domain containing protein-3; LATS1, large tumor suppressor-1; MST1, mammalian Ste20-like kinase 1; YAP, yes-associated protein; TEAD, TEA domain transcription factors; RT-qPCR, reverse transcription quantitative polymerase chain reaction; NSCLC, non-small cell lung cancer; CTGF, connective tissue growth factor; Cyr61, Cysteine-rich angiogenic inducer 61; SPSS, statistical program for social sciences; MTT, methylthiazolyl diphenyl tetrazolium bromide; TNM stage, Tumor, Node, Metastasis stage; PDZ domain, PSD95/DLG/ZO1 domain; Fat, Fat atypical-cadherin; NF2, neurofibromin 2; AMOT, angiomotin; Wnt pathway, Wingless/Integrated pathway; DVL, dishevelled segment polarity protein; LATS1, large tumor suppressor kinase-1; Yki, yorkie; ECL, enhanced chemiluminescence; PVDF membrane, Polyvinylidene Fluoride membrane; GFP, Green Fluorescent protein; TCGA database, The Cancer Genome Atlas database; FRMD6, FERM domain containing 6; HBE, normal human bronchial epithelial cell line; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; EV, empty vector; NC, negative control.
Acknowledgments
The work was supported by grants from the National Natural Science Foundation of China (No. 81572854 and No. 81772489 to Enhua Wang, No. 81401885 to Xuyong Lin, No. 81672302 to Di Zhang) and China Postdoctoral Science Foudation (No. 2018M641737 to Qiang Han).
Disclosure
All authors declare that they have no conflicts of interest in this work.
References
1. Harvey K, Tapon N. The Salvador-Warts-Hippo pathway-an emerging tumor-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi:10.1038/nrc2070
2. Hu C, Sun J, Du J, et al. The Hippo-YAP pathway regulates the proliferation of alveolar epithelial progenitors after acute lung injury. Cell Biol Int. 2019 [Epub ahead of print]. doi:10.1002/cbin.11098
3. Gao L, Cao H, Cheng X. A positive feedback regulation between long noncoding RNA SNHG1 and YAP1 modulates growth and metastasis in laryngeal squamous cell carcinoma. Am J Cnacer Res. 2018;8:1712–1724.
4. Lehmann W, Mossmann D, KIeemann J, et al. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun. 2016;7:10498. doi:10.1038/ncomms10498
5. Song GQ, Zhao Y. MAC30 knockdown involved in the activation of the hippo signaling pathway in breast cancer cells. Biol Chem. 2018 [Epub ahead of print]. doi:10.1515/hsz-2018-0250
6. Martin D, Degese MS, Vitale-Cross L, et al. Assembly and activation of the hippo signalome by FAT1 tumor suppressor. Nat Commun. 2018;9:2372. doi:10.1038/s41467-018-04590-1
7. Plouffe SW, Lin KC, Moore JL, et al. The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J Bio Chem. 2018;193:11230–11240. doi:10.1074/jbc.RA118.002715
8. Kremerskothen J, Plaas C, Buther K, et al. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun. 2003;300:862–867.
9. Han Q, Kremerskothen J, Lin X, et al. WWC3 inhibits epithelial-mesenchymal transition of lung cancer by activating Hippo-YAP signaling. Onco Targets Ther. 2017;10:2931–2942. doi:10.2147/OTT.S124790
10. Han Q, Lin X, Zhang X, et al. WWC3 regulates the Wnt and hippo pathways via dishevelled proteins and large tumor suppressor 1. To suppress lung cancer invasion and metastasis. J Pathol. 2017;242:435–447. doi:10.1002/path.4919
11. Tate G, Kishimoto K, Mitsuya T. A novel mutation of the FAT2 gene in spinal meningioma. Oncol Lett. 2016;12:3393–3396. doi:10.3892/ol.2016.5063
12. Pan Z, Shang Y, Jia M, et al. Structural and biochemical characterization of the interaction between LGN and FRMPD1. J Mol Bio. 2013;425:1039–1049. doi:10.1016/j.jmb.2013.01.003
13. An N, Blummer JB, Bernard ML, Lanier SM. The PDZ and bind 4.1 containing protein FRMPD1 regulates the subcellular location of activator of G-protein signaling 3 and its interaction with G-proteins. J Biol Chem. 2008;283:24718–24728.
14. McCartney BM, Kuikauskas RM, LaJeunesse DR, Fehon RG. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324.
15. Cox AG, Tsomides A, Yimlamai D, et al. Yap regulates glucose utilization and sustains nucleotide synthesis to enable organ growth. EMBO J. 2018;37:
16. Moleirinhol S, Tilston-Lunel A, Angus L, Gunn-Moore F, Reynolds PA. The expending family of FERM proteins. Biochem J. 2013;452:183–193. doi:10.1042/BJ20121642
17. Angus L, Molerinhol S, Herron L, et al. Willin/FRMPD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene. 2012;31:238–250. doi:10.1038/onc.2011.224
18. Travis WD, Brambilla E, Burke AP, et al. The 2015 World Health Organization classification of lung tumor: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi:10.1097/JTO.0000000000000630
19. Goldstraw P. Updated staging system for lung cancer. Surg Oncol Clin North Am. 2011;20:655–666. doi:10.1016/j.soc.2011.07.005
20. Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. A molecular mechanism that links hippo signaling to the inhibition of Wnt/β-catenin signaling. EMBO J. 2012;31:1109–1122. doi:10.1038/emboj.2011.487
21. Lin XY, Zhang XP, Wu JH, et al. Expression of LATS1 contributes to good prognosis and can negatively regulate YAP oncoprotein in non-small-lung-cancer. Tumour Biol. 2014;35:6435–6443. doi:10.1007/s13277-014-1826-z
22. Testa U, Casterlli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers (Basel). 2018;10:248. doi:10.3390/cancers10110400
23. Bircan HA, Gurbuz N, Patear A, et al. Elongation factor-2 kinase (eEF-2K) expression is associated with poor patient survival and promotes proliferation, invasion and tumor growth of lung cancer. Lung Cancer. 2018;124:31–39. doi:10.1016/j.lungcan.2018.07.027
24. Moroishi T, Hayashi T, Pan WW, et al. The Hippo pathway kinase LATS1/2 suppress cancer immunity. Cell. 2016;167:1525–1539. doi:10.1016/j.cell.2016.11.005
25. James van Rensburg HJ, Azad T, Ling M, et al. The Hippo pathway component TAZ promotes immune evasion in human cancer through PD-L1. Cancer Res. 2018;78:1457–1470. doi:10.1158/0008-5472.CAN-17-3139
26. Hsu PC, Yang CT, Jablons DM, You L. The role of yes-associated protein (YAP) in regulating programmed death –ligand 1 (PD-L1) in thoracic cancer. Biomedicines. 2018.pii:E114. doi:10.3390/biomedicines6040114
Supplementary Materials
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.